Protein Name: Pyruvate dehydrogenase E1 component subunit alpha, somatic form, mitochondrial
|
UniprotKB/SwissProt ID: ODPA_MOUSE (P35486)
Gene Name: Pdha1
Synonyms: Pdha-1
Organism: Mus musculus (Mouse).
Function: The pyruvate dehydrogenase complex catalyzes the overall conversion of pyruvate to acetyl-CoA and CO(2), and thereby links the glycolytic pathway to the tricarboxylic cycle.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Mitochondrion matrix.
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 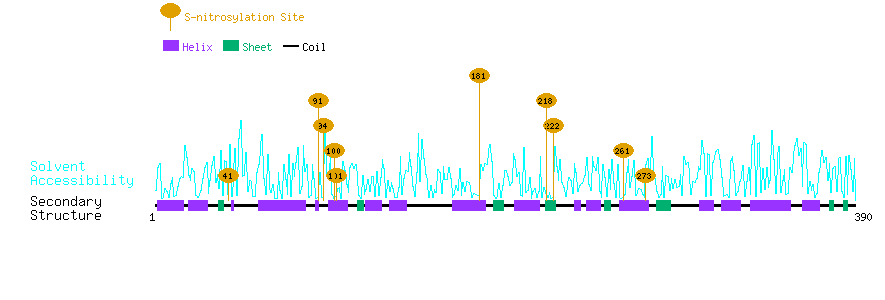 |
|

