Protein Name: Cellular tumor antigen p53
|
UniprotKB/SwissProt ID: P53_HUMAN (P04637)
Gene Name: TP53
Synonyms: P53
Organism: Homo sapiens (Human).
Function: Acts as a tumor suppressor in many tumor types; induces growth arrest or apoptosis depending on the physiological circumstances and cell type. Involved in cell cycle regulation as a trans-activator that acts to negatively regulate cell division by controlling a set of genes required for this process. One of the activated genes is an inhibitor of cyclin-dependent kinases. Apoptosis induction seems to be mediated either by stimulation of BAX and FAS antigen expression, or by repression of Bcl-2 expression. In cooperation with mitochondrial PPIF is involved in activating oxidative stress-induced necrosis; the function is largely independent of transcription. Induces the transcription of long intergenic non-coding RNA p21 (lincRNA-p21) and lincRNA- Mkln1. LincRNA-p21 participates in TP53-dependent transcriptional repression leading to apoptosis and seem to have to effect on cell-cycle regulation. Implicated in Notch signaling cross-over. Prevents CDK7 kinase activity when associated to CAK complex in response to DNA damage, thus stopping cell cycle progression. Isoform 2 enhances the transactivation activity of isoform 1 from some but not all TP53-inducible promoters. Isoform 4 suppresses transactivation activity and impairs growth suppression mediated by isoform 1. Isoform 7 inhibits isoform 1-mediated apoptosis.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cytoplasm. Nucleus. Nucleus, PML body. Endoplasmic reticulum. Mitochondrion matrix. Note=Interaction with BANP promotes nuclear localization. Recruited into PML bodies together with CHEK2. Translocates to mitochondria upon oxidative stress. Isoform 1: Nu
|
PDB :
( If your security settings prevent Jmol from running, please register http://140.138.144.145/ as a safe location in your Java settings. ) |
|
Protein disease:
|
| Disease database |
Database Entry |
Disease information | | KEGG | H00020 | Colorectal cancer | | HPRD | 01859 | Adrenocortical carcinoma, pediatric | | HPRD | 01859 | Breast cancer | | HPRD | 01859 | Choroid plexus papilloma | | HPRD | 01859 | Colon tumors, concurrent multiple primary | | HPRD | 01859 | Hepatoblastoma | | HPRD | 01859 | Hepatocellular carcinoma | | HPRD | 01859 | Histiocytoma, malignant fibrous | | HPRD | 01859 | Li-Fraumeni syndrome | | HPRD | 01859 | Li-Fraumeni syndrome 1 | | HPRD | 01859 | Multiple malignancy syndrome | | HPRD | 01859 | Nasopharyngeal carcinoma | | HPRD | 01859 | Osteosarcoma | | HPRD | 01859 | Pancreatic cancer | | HPRD | 01859 | Tp53 polymorphism |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 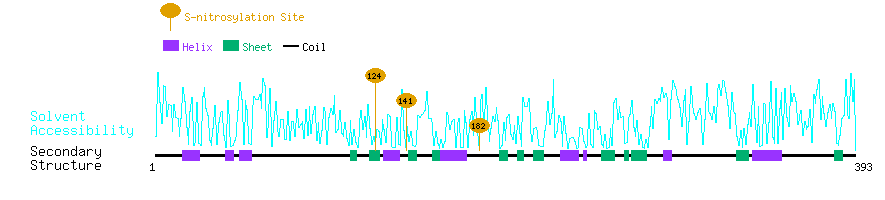 |
|

