Protein Name: Protein DJ-1
|
UniprotKB/SwissProt ID: PARK7_HUMAN (Q99497)
Gene Name: PARK7
Organism: Homo sapiens (Human).
Function: Protects cells against oxidative stress and cell death. Plays a role in regulating expression or stability of the mitochondrial uncoupling proteins SLC25A14 and SLC25A27 in dopaminergic neurons of the substantia nigra pars compacta and attenuates the oxidative stress induced by calcium entry into the neurons via L-type channels during pacemaking. Eliminates hydrogen peroxide and protects cells against hydrogen peroxide-induced cell death. May act as an atypical peroxiredoxin-like peroxidase that scavenges hydrogen peroxide. Following removal of a C-terminal peptide, displays protease activity and enhanced cytoprotective action against oxidative stress-induced apoptosis. Stabilizes NFE2L2 by preventing its association with KEAP1 and its subsequent ubiquitination. Binds to OTUD7B and inhibits its deubiquitinating activity. Enhances RELA nuclear translocation. Binds to a number of mRNAs containing multiple copies of GG or CC motifs and partially inhibits their translation but dissociates following oxidative stress. Required for correct mitochondrial morphology and function and for autophagy of dysfunctional mitochondria. Regulates astrocyte inflammatory responses. Acts as a positive regulator of androgen receptor-dependent transcription. Prevents aggregation of SNCA. Plays a role in fertilization. Has no proteolytic activity. Has cell-growth promoting activity and transforming activity. May function as a redox-sensitive chaperone. May regulate lipid rafts-dependent endocytosis in astrocytes and neuronal cells.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cell membrane; Lipid-anchor. Cytoplasm. Nucleus. Membrane raft. Mitochondrion. Note=Under normal conditions, located predominantly in the cytoplasm and, to a lesser extent, in the nucleus and mitochondrion. Translocates to the mitochondrion and subsequen
|
PDB :
( If your security settings prevent Jmol from running, please register http://140.138.144.145/ as a safe location in your Java settings. ) |
|
Protein disease:
|
| Disease database |
Database Entry |
Disease information | | HPRD | 03961 | Amyotrophic lateral sclerosis-parkinsonism/dementia complex 2 | | HPRD | 03961 | Parkinson disease 7, autosomal recessive early-onset |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 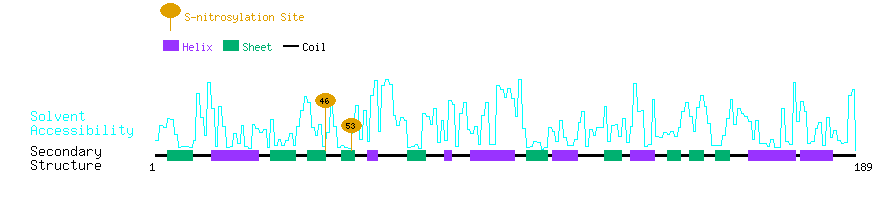 |
|
