Protein Name: [Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 2, mitochondrial
|
UniprotKB/SwissProt ID: PDK2_MOUSE (Q9JK42)
Gene Name: Pdk2
Organism: Mus musculus (Mouse).
Function: Serine/threonine kinase that plays a key role in the regulation of glucose and fatty acid metabolism and homeostasis via phosphorylation of the pyruvate dehydrogenase subunits PDHA1 and PDHA2. This inhibits pyruvate dehydrogenase activity, and thereby regulates metabolite flux through the tricarboxylic acid cycle, down-regulates aerobic respiration and inhibits the formation of acetyl-coenzyme A from pyruvate. Inhibition of pyruvate dehydrogenase decreases glucose utilization and increases fat metabolism. Mediates cellular responses to insulin. Plays an important role in maintaining normal blood glucose levels and in metabolic adaptation to nutrient availability. Via its regulation of pyruvate dehydrogenase activity, plays an important role in maintaining normal blood pH and in preventing the accumulation of ketone bodies under starvation. Plays a role in the regulation of cell proliferation and in resistance to apoptosis under oxidative stress. Plays a role in p53/TP53-mediated apoptosis.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Mitochondrion matrix.
| |
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 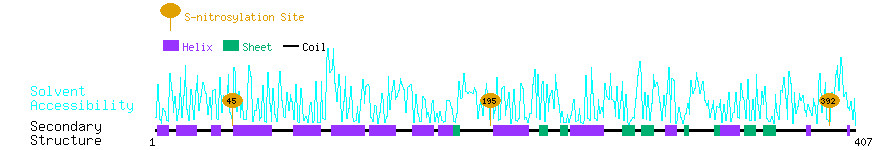 |
|

