Protein Name: Eosinophil peroxidase
|
UniprotKB/SwissProt ID: PERE_HUMAN (P11678)
Gene Name: EPX
Synonyms: EPER, EPO, EPP
Organism: Homo sapiens (Human).
Function: Mediates tyrosine nitration of secondary granule proteins in mature resting eosinophils. Shows significant inhibitory activity towards Mycobacterium tuberculosis H37Rv by inducing bacterial fragmentation and lysis.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cytoplasmic granule. Note=Cytoplasmic granules of eosinophils.
| |
|
Protein disease:
|
| Disease database |
Database Entry |
Disease information | | HPRD | 00576 | Eosinophil peroxidase deficiency |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 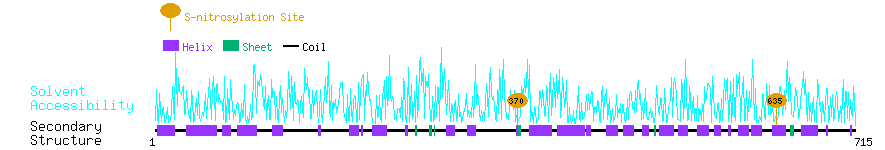 |
|

