Protein Name: Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1
|
UniprotKB/SwissProt ID: PIN1_MOUSE (Q9QUR7)
Gene Name: Pin1
Organism: Mus musculus (Mouse).
Function: Essential PPIase that regulates mitosis presumably by interacting with NIMA and attenuating its mitosis-promoting activity. Displays a preference for an acidic residue N-terminal to the isomerized proline bond. Catalyzes pSer/Thr-Pro cis/trans isomerizations. Down-regulates kinase activity of BTK. Can transactivate multiple oncogenes and induce centrosome amplification, chromosome instability and cell transformation (By similarity). Required for the efficient dephosphorylation and recycling of RAF1 after mitogen activation (By similarity). Binds and targets BCL6 and PML for degradation in a phosphorylation- dependent manner.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Nucleus (By similarity). Nucleus speckle (By similarity). Cytoplasm (By similarity). Note=Colocalizes with NEK6 in the nucleus. Mainly localized in the nucleus but phosphorylation at Ser-73 by DAPK1 results in inhibition of its nuclear localization (By s
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 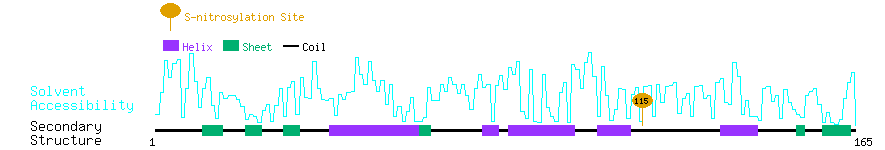 |
|

