Protein Name: Serine/threonine-protein phosphatase 2B catalytic subunit alpha isoform
|
UniprotKB/SwissProt ID: PP2BA_RAT (P63329)
Gene Name: Ppp3ca
Synonyms: Calna
Organism: Rattus norvegicus (Rat).
Function: Calcium-dependent, calmodulin-stimulated protein phosphatase. Many of the substrates contain a PxIxIT motif. This subunit may have a role in the calmodulin activation of calcineurin. Dephosphorylates DNM1L, HSPB1 and SSH1 (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cell membrane (By similarity). Cell membrane, sarcolemma (By similarity). Nucleus (By similarity). Note=Colocalizes with ACTN1 and MYOZ2 at the Z line in heart and skeletal muscle (By similarity).
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 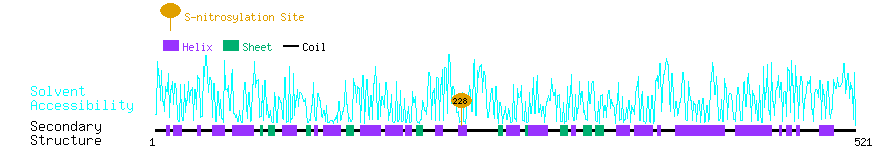 |
|
