Protein Name: Peptidyl-prolyl cis-trans isomerase D
|
UniprotKB/SwissProt ID: PPID_HUMAN (Q08752)
Gene Name: PPID
Synonyms: CYP40, CYPD
Organism: Homo sapiens (Human).
Function: PPIases accelerate the folding of proteins. It catalyzes the cis-trans isomerization of proline imidic peptide bonds in oligopeptides. Proposed to act as a co-chaperone in HSP90 complexes such as in unligated steroid receptors heterocomplexes. Different co-chaperones seem to compete for association with HSP90 thus establishing distinct HSP90-co-chaperone-receptor complexes with the potential to exert tissue-specific receptor activity control. May have a preference for estrogen receptor complexes and is not found in glucocorticoid receptor complexes. May be involved in cytoplasmic dynein-dependent movement of the receptor from the cytoplasm to the nucleus. May regulate MYB by inhibiting its DNA- binding activity. Involved in regulation of AHR signaling by promoting the formation of the AHR:ARNT dimer; the function is independent of HSP90 but requires the chaperone activity. Involved in regulation of UV radiation-induced apoptosis. Promotes cell viability in anaplastic lymphoma kinase-positive anaplastic large- cell lymphoma (ALK+ ALCL) cell lines. May be involved in hepatitis C virus (HCV) replication and release.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cytoplasm. Nucleus, nucleolus. Nucleus, nucleoplasm.
| |
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 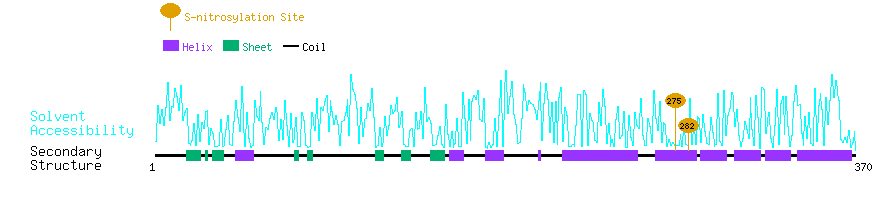 |
|

