Protein Name: Glycogen phosphorylase, brain form
|
UniprotKB/SwissProt ID: PYGB_MOUSE (Q8CI94)
Gene Name: Pygb
Organism: Mus musculus (Mouse).
Function: Phosphorylase is an important allosteric enzyme in carbohydrate metabolism. Enzymes from different sources differ in their regulatory mechanisms and in their natural substrates. However, all known phosphorylases share catalytic and structural properties (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization:
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 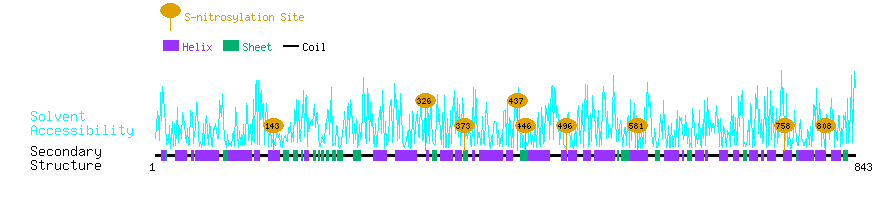 |
|

