Protein Name: Ras-related C3 botulinum toxin substrate 2
|
UniprotKB/SwissProt ID: RAC2_HUMAN (P15153)
Gene Name: RAC2
Organism: Homo sapiens (Human).
Function: Plasma membrane-associated small GTPase which cycles between an active GTP-bound and inactive GDP-bound state. In active state binds to a variety of effector proteins to regulate cellular responses, such as secretory processes, phagocytose of apoptotic cells and epithelial cell polarization. Augments the production of reactive oxygen species (ROS) by NADPH oxidase.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cytoplasm. Note=Membrane-associated when activated.
|
PDB :
( If your security settings prevent Jmol from running, please register http://140.138.144.145/ as a safe location in your Java settings. ) |
|
Protein disease:
|
| Disease database |
Database Entry |
Disease information | | KEGG | H00020 | Colorectal cancer | | HPRD | 03628 | Neutrophil immunodeficiency syndrome |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 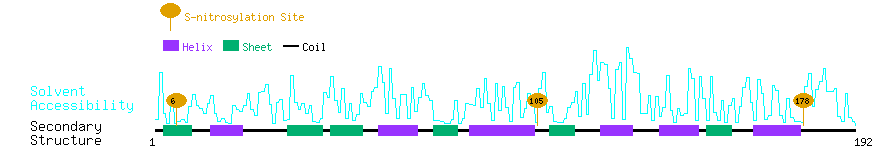 |
|

