Protein Name: GTPase NRas
|
UniprotKB/SwissProt ID: RASN_HUMAN (P01111)
Gene Name: NRAS
Synonyms: HRAS1
Organism: Homo sapiens (Human).
Function: Ras proteins bind GDP/GTP and possess intrinsic GTPase activity.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cell membrane; Lipid-anchor; Cytoplasmic side. Golgi apparatus membrane; Lipid-anchor. Note=Shuttles between the plasma membrane and the Golgi apparatus.
|
PDB :
( If your security settings prevent Jmol from running, please register http://140.138.144.145/ as a safe location in your Java settings. ) |
|
Protein disease:
|
| Disease database |
Database Entry |
Disease information | | KEGG | H00003 | Acute myeloid leukemia (AML) | | KEGG | H00010 | Multiple myeloma | | KEGG | H00016 | Oral cancer | | KEGG | H00032 | Thyroid cancer | | KEGG | H00033 | Adrenal carcinoma | | KEGG | H00038 | Malignant melanoma | | KEGG | H00048 | Hepatocellular carcinoma | | KEGG | H00108 | Autoimmune lymphoproliferative syndromes (ALPS), including the following five diseases: CD95 (Fas) d | | KEGG | H00523 | Noonan syndrome and related disorders, including: Noonan syndrome (NS); Leopard syndrome (LS); Noona | | OMIM | 114500 | COLORECTAL CANCER; CRC ;;COLON CANCER | | OMIM | 162900 | NEVUS, EPIDERMAL ;;PIGMENTED MOLES;; NEVUS, KERATINOCYTIC, NONEPIDERMOLYTIC | | OMIM | 164790 | NEUROBLASTOMA RAS VIRAL ONCOGENE HOMOLOG; NRAS ;;ONCOGENE NRAS; NRAS1 AUTOIMMUNE LYMPHOPROLI | | OMIM | 188470 | THYROID CARCINOMA, FOLLICULAR; FTC | | OMIM | 613224 | NOONAN SYNDROME 6; NS6 | | OMIM | 614470 | AUTOIMMUNE LYMPHOPROLIFERATIVE SYNDROME, TYPE IV; ALPS4 | | HPRD | 01273 | Colorectal cancer | | HPRD | 01273 | Thyroid carcinoma, follicular |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 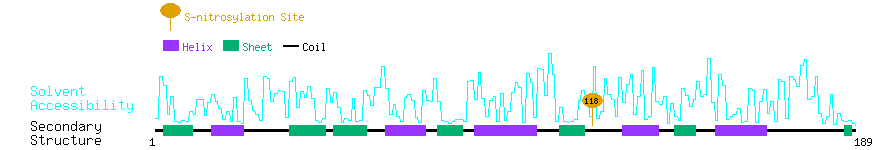 |
|
