Protein Name: Transforming protein RhoA
|
UniprotKB/SwissProt ID: RHOA_MOUSE (Q9QUI0)
Gene Name: Rhoa
Synonyms: Arha, Arha2
Organism: Mus musculus (Mouse).
Function: Regulates a signal transduction pathway linking plasma membrane receptors to the assembly of focal adhesions and actin stress fibers. Involved in a microtubule-dependent signal that is required for the myosin contractile ring formation during cell cycle cytokinesis. Plays an essential role in cleavage furrow formation. May be an activator of PLCE1. Activated by ARHGEF2, which promotes the exchange of GDP for GTP. Essential for the SPATA13-mediated regulation of cell migration and adhesion assembly and disassembly. The MEMO1-RHOA-DIAPH1 signaling pathway plays an important role in ERBB2-dependent stabilization of microtubules at the cell cortex. It controls the localization of APC and CLASP2 to the cell membrane, via the regulation of GSK3B activity. In turn, membrane-bound APC allows the localization of the MACF1 to the cell membrane, which is required for microtubule capture and stabilization (By similarity). Required for the apical junction formation of keratinocyte cell-cell adhesion.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cell membrane; Lipid-anchor; Cytoplasmic side (By similarity). Cytoplasm, cytoskeleton (By similarity). Cleavage furrow (By similarity). Cytoplasm, cell cortex (By similarity). Midbody (By similarity). Cell projection, lamellipodium. Note=Translocates to
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 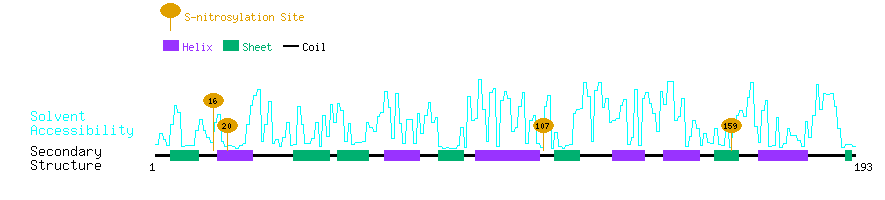 |
|
