Protein Name: 60S ribosomal protein L12
|
UniprotKB/SwissProt ID: RL12_MOUSE (P35979)
Gene Name: Rpl12
Organism: Mus musculus (Mouse).
Function: Binds directly to 26S ribosomal RNA.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization:
|
PDB :
( If your security settings prevent Jmol from running, please register http://140.138.144.145/ as a safe location in your Java settings. ) |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 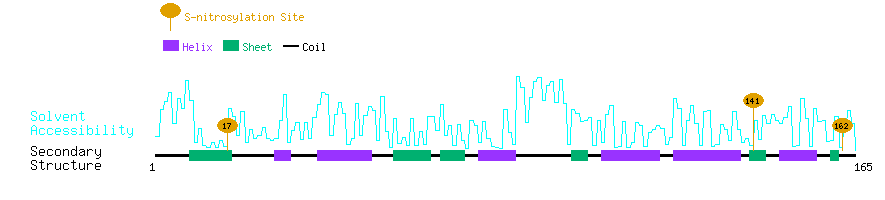 |
|
