Protein Name: 60S ribosomal protein L5
|
UniprotKB/SwissProt ID: RL5_HUMAN (P46777)
Gene Name: RPL5
Organism: Homo sapiens (Human).
Function: Required for rRNA maturation and formation of the 60S ribosomal subunits. This protein binds 5S RNA.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cytoplasm. Nucleus, nucleolus.
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 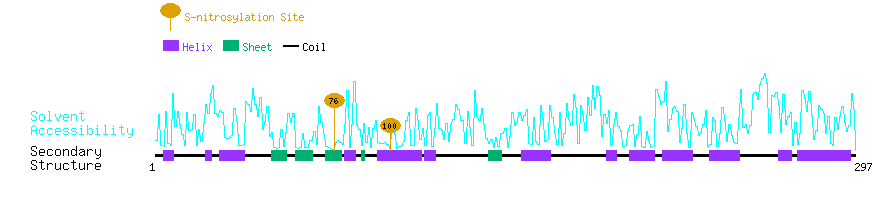 |
|
