Protein Name: Ras-related protein R-Ras2
|
UniprotKB/SwissProt ID: RRAS2_HUMAN (P62070)
Gene Name: RRAS2
Synonyms: TC21
Organism: Homo sapiens (Human).
Function: It is a plasma membrane-associated GTP-binding protein with GTPase activity. Might transduce growth inhibitory signals across the cell membrane, exerting its effect through an effector shared with the Ras proteins but in an antagonistic fashion.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cell membrane; Lipid-anchor; Cytoplasmic side (By similarity). Note=Inner surface of plasma membrane possibly with attachment requiring acylation of the C-terminal cysteine (By similarity with RAS).
|
PDB :
( If your security settings prevent Jmol from running, please register http://140.138.144.145/ as a safe location in your Java settings. ) |
|
Protein disease:
|
| Disease database |
Database Entry |
Disease information | | OMIM | 600098 | RELATED RAS VIRAL ONCOGENE HOMOLOG 2; RRAS2 ;;ONCOGENE RRAS2;; TERATOCARCINOMA ONCOGENE TC21 | | HPRD | 02518 | Ovarian carcinoma |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 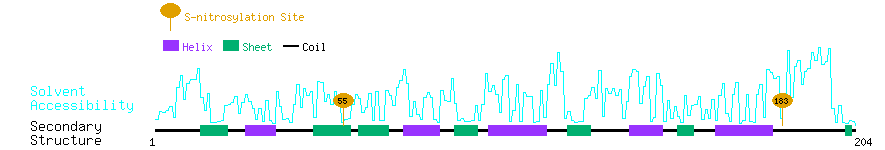 |
|
