Protein Name: Selenium-binding protein 1
|
UniprotKB/SwissProt ID: SBP1_MOUSE (P17563)
Gene Name: Selenbp1
Synonyms: Lpsb
Organism: Mus musculus (Mouse).
Function: Selenium-binding protein which may be involved in the sensing of reactive xenobiotics in the cytoplasm. May be involved in intra-Golgi protein transport (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Nucleus (By similarity). Cytoplasm, cytosol. Membrane; Peripheral membrane protein (By similarity). Note=May associate with Golgi membrane. May associate with the membrane of autophagosomes (By similarity).
| |
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 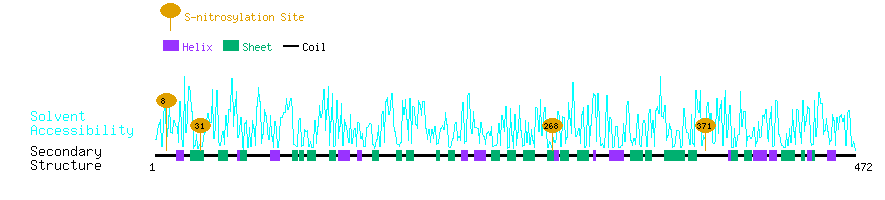 |
|

