Protein Name: Serine/arginine-rich splicing factor 7
|
UniprotKB/SwissProt ID: SRSF7_HUMAN (Q16629)
Gene Name: SRSF7
Synonyms: SFRS7
Organism: Homo sapiens (Human).
Function: Required for pre-mRNA splicing. Can also modulate alternative splicing in vitro. Represses the splicing of MAPT/Tau exon 10. May function as export adapter involved in mRNA nuclear export such as of histone H2A. Binds mRNA which is thought to be transferred to the NXF1-NXT1 heterodimer for export (TAP/NXF1 pathway); enhances NXF1-NXT1 RNA-binding activity. RNA-binding is semi-sequence specific.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Nucleus. Cytoplasm.
|
PDB :
( If your security settings prevent Jmol from running, please register http://140.138.144.145/ as a safe location in your Java settings. ) |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 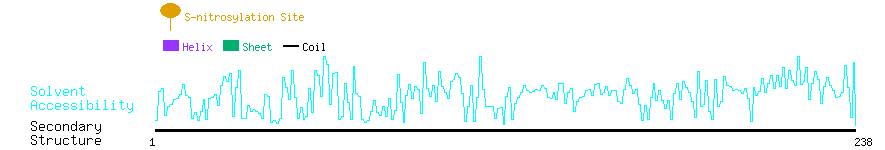 |
|

