Protein Name: Serine/arginine-rich splicing factor 9
|
UniprotKB/SwissProt ID: SRSF9_HUMAN (Q13242)
Gene Name: SRSF9
Synonyms: SFRS9, SRP30C
Organism: Homo sapiens (Human).
Function: Plays a role in constitutive splicing and can modulate the selection of alternative splice sites. Represses the splicing of MAPT/Tau exon 10.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Nucleus. Note=Cellular stresses such as heat shock may induce localization to discrete nuclear bodies termed SAM68 nuclear bodies (SNBs), HAP bodies, or stress bodies. Numerous splicing factors including SRSF1/SFRS1/SF2, SRSF7/SFRS7, SAFB and KHDRBS1/SAM
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 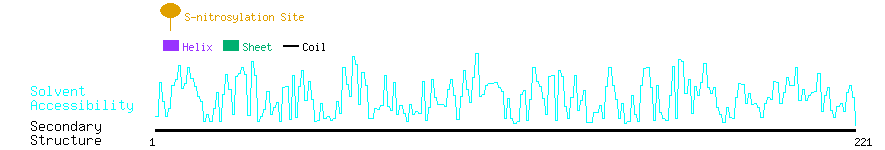 |
|

