Protein Name: Succinyl-CoA ligase [ADP-forming] subunit beta, mitochondrial
|
UniprotKB/SwissProt ID: SUCB1_MOUSE (Q9Z2I9)
Gene Name: Sucla2
Organism: Mus musculus (Mouse).
Function: Catalyzes the ATP-dependent ligation of succinate and CoA to form succinyl-CoA (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Mitochondrion.
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 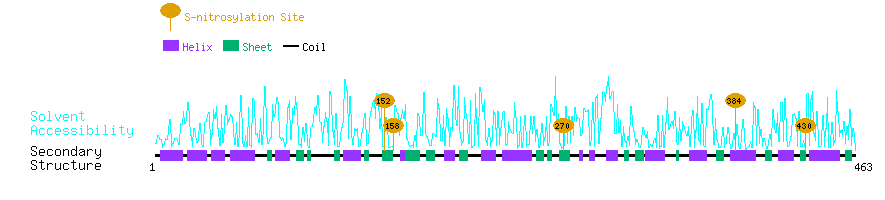 |
|

