Protein Name: Surfeit locus protein 4
|
UniprotKB/SwissProt ID: SURF4_MOUSE (Q64310)
Gene Name: Surf4
Synonyms: Surf-4
Organism: Mus musculus (Mouse).
Function: May play a role in the maintenance of the architecture of the endoplasmic reticulum-Golgi intermediate compartment and of the Golgi (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Endoplasmic reticulum membrane; Multi-pass membrane protein (By similarity). Endoplasmic reticulum-Golgi intermediate compartment membrane; Multi-pass membrane protein (By similarity). Golgi apparatus membrane; Multi-pass membrane protein (By similarity)
| |
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 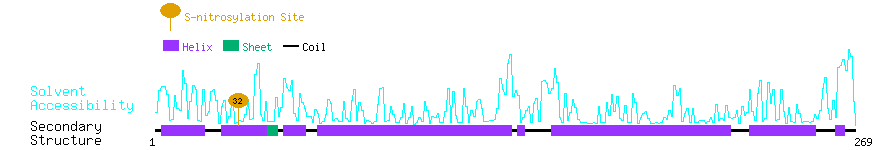 |
|

