Protein Name: Tubulin alpha-4A chain
|
UniprotKB/SwissProt ID: TBA4A_MOUSE (P68368)
Gene Name: Tuba4a
Synonyms: Tuba4
Organism: Mus musculus (Mouse).
Function: Tubulin is the major constituent of microtubules. It binds two moles of GTP, one at an exchangeable site on the beta chain and one at a non-exchangeable site on the alpha chain.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cytoplasm, cytoskeleton.
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 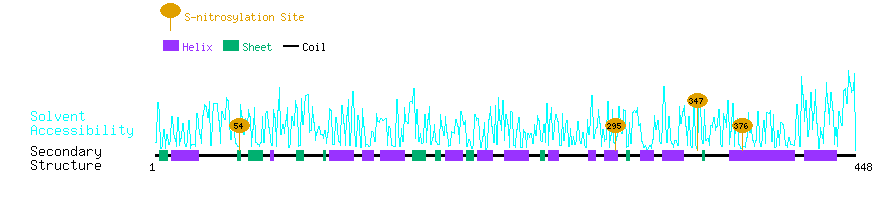 |
|

