Protein Name: 3-ketoacyl-CoA thiolase, mitochondrial
|
UniprotKB/SwissProt ID: THIM_MOUSE (Q8BWT1)
Gene Name: Acaa2
Organism: Mus musculus (Mouse).
Function: Abolishes BNIP3-mediated apoptosis and mitochondrial damage (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Mitochondrion (By similarity). Note=Colocalizes with BNIP3 in the mitochondria (By similarity).
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 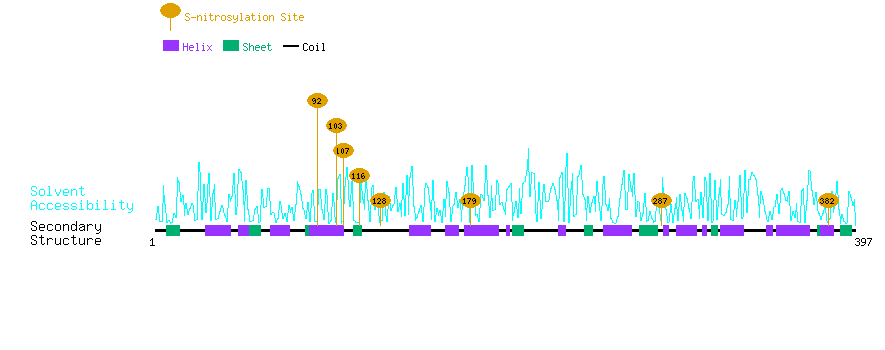 |
|

