Protein Name: Heat shock protein 75 kDa, mitochondrial
|
UniprotKB/SwissProt ID: TRAP1_MOUSE (Q9CQN1)
Gene Name: Trap1
Synonyms: Hsp75
Organism: Mus musculus (Mouse).
Function: Chaperone that expresses an ATPase activity (By similarity). Involved in maintaining mitochondrial function and polarization, most likely through stabilization of mitochondrial complex I (By similarity). Is a negative regulator of mitochondrial respiration able to modulate the balance between oxidative phosphorylation and aerobic glycolysis (By similarity). The impact of TRAP1 on mitochondrial respiration is probably mediated by modulation of mitochondrial SRC and inhibition of SDHA (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Mitochondrion (By similarity). Mitochondrion inner membrane (By similarity). Mitochondrion matrix (By similarity).
| |
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 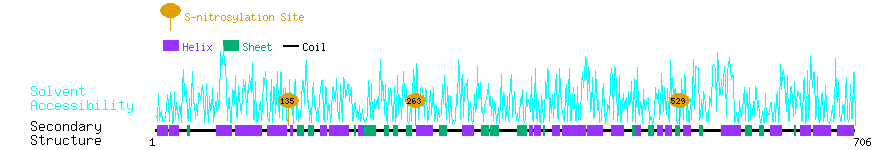 |
|

