Protein Name: NEDD8-activating enzyme E1 regulatory subunit
|
UniprotKB/SwissProt ID: ULA1_MOUSE (Q8VBW6)
Gene Name: Nae1
Synonyms: Appbp1
Organism: Mus musculus (Mouse).
Function: Regulatory subunit of the dimeric UBA3-NAE1 E1 enzyme. E1 activates NEDD8 by first adenylating its C-terminal glycine residue with ATP, thereafter linking this residue to the side chain of the catalytic cysteine, yielding a NEDD8-UBA3 thioester and free AMP. E1 finally transfers NEDD8 to the catalytic cysteine of UBE2M. Necessary for cell cycle progression through the S-M checkpoint. Overexpression of NAE1 causes apoptosis through deregulation of NEDD8 conjugation (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cell membrane (By similarity). Note=Colocalizes with APP in lipid rafts (By similarity).
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 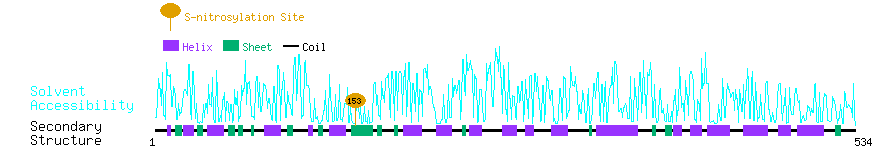 |
|

