Protein Name: V-type proton ATPase subunit B, brain isoform
|
UniprotKB/SwissProt ID: VATB2_HUMAN (P21281)
Gene Name: ATP6V1B2
Synonyms: ATP6B2, VPP3
Organism: Homo sapiens (Human).
Function: Non-catalytic subunit of the peripheral V1 complex of vacuolar ATPase. V-ATPase is responsible for acidifying a variety of intracellular compartments in eukaryotic cells.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Endomembrane system; Peripheral membrane protein. Melanosome. Note=Endomembrane. Identified by mass spectrometry in melanosome fractions from stage I to stage IV.
| |
|
Protein disease:
|
| Disease database |
Database Entry |
Disease information | | OMIM | 606939 | ATPasE, H TRANSPORTING, LYSOSOMAL, 56/58-KD, V1 SUBUNIT B, ISOFORM 2; ATP6V1B2 ;;ATP6B2;; V |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 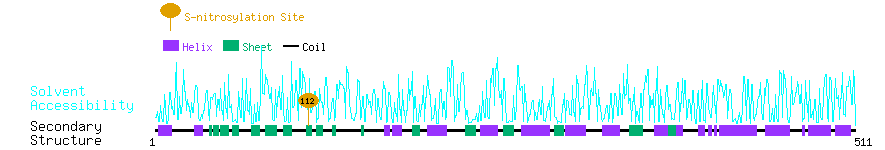 |
|

