Protein Name: V-type proton ATPase subunit H
|
UniprotKB/SwissProt ID: VATH_MOUSE (Q8BVE3)
Gene Name: Atp6v1h
Organism: Mus musculus (Mouse).
Function: Subunit of the peripheral V1 complex of vacuolar ATPase. Subunit H activates the ATPase activity of the enzyme and couples ATPase activity to proton flow. Vacuolar ATPase is responsible for acidifying a variety of intracellular compartments in eukaryotic cells, thus providing most of the energy required for transport processes in the vacuolar system. Involved in the endocytosis mediated by clathrin-coated pits, required for the formation of endosomes (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization:
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 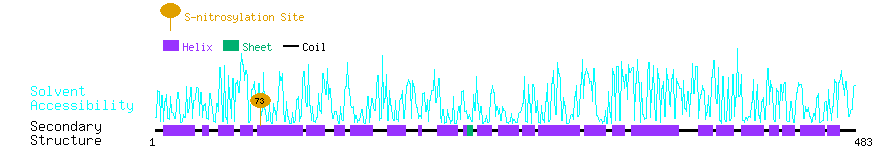 |
|

