Protein Name: Voltage-dependent anion-selective channel protein 1
|
UniprotKB/SwissProt ID: VDAC1_MOUSE (Q60932)
Gene Name: Vdac1
Synonyms: Vdac5
Organism: Mus musculus (Mouse).
Function: Forms a channel through the mitochondrial outer membrane and also the plasma membrane. The channel at the outer mitochondrial membrane allows diffusion of small hydrophilic molecules; in the plasma membrane it is involved in cell volume regulation and apoptosis. It adopts an open conformation at low or zero membrane potential and a closed conformation at potentials above 30-40 mV. The open state has a weak anion selectivity whereas the closed state is cation-selective. May participate in the formation of the permeability transition pore complex (PTPC) responsible for the release of mitochondrial products that triggers apoptosis.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Isoform Mt-VDAC1: Mitochondrion outer membrane; Multi-pass membrane protein. Isoform Pl-VDAC1: Cell membrane; Multi-pass membrane protein.
|
PDB :
( If your security settings prevent Jmol from running, please register http://140.138.144.145/ as a safe location in your Java settings. ) |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 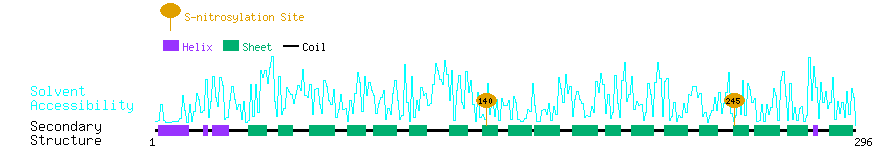 |
|
