Protein Name: Voltage-dependent anion-selective channel protein 2
|
UniprotKB/SwissProt ID: VDAC2_MOUSE (Q60930)
Gene Name: Vdac2
Synonyms: Vdac6
Organism: Mus musculus (Mouse).
Function: Forms a channel through the mitochondrial outer membrane that allows diffusion of small hydrophilic molecules. The channel adopts an open conformation at low or zero membrane potential and a closed conformation at potentials above 30-40 mV. The open state has a weak anion selectivity whereas the closed state is cation- selective (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Mitochondrion outer membrane.
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 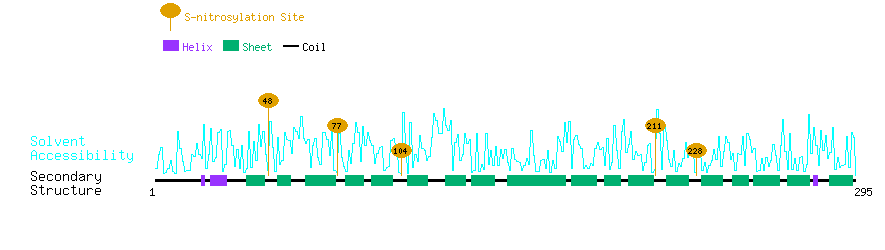 |
|

