About PhosK 3D
Abstract
|
With the high-throughput of mass spectrometry-based phospho-proteomics, the desire to comprehensively annotate the catalytic kinases for in vivo phosphorylation sites has motivated. Herein, a new web resource (PhosK3D) has been developed for performing a large-scale prediction of over 100 kinase-specific groups which contain sufficient number of experimental substrate sites. In an attempt to physically investigate the phosphorylation sites on protein tertiary structures, all experimental phosphorylation sites are mapped to the protein entries of Protein Data Bank by sequence identity. It resulted in a total of 4508 phosphorylation sites containing the protein three-dimensional (3D) structures.To identify phosphorylation sites on protein 3D structures, this work incorporated support vector machine (SVM) with the information of spatial amino acid composition and structural alphabet, which is a new feature to encode a 3D structure fragment of protein backbones into 23 structural alphabets. After the cross-validation evaluation, most of the kinase-specific models trained with the consideration of structural information outperform the models considering only the sequence information. Moreover, the independent testing set which is not included in the training set has demonstrated that the KinasePhos3D could provide a comparable performance to other popular tools. This work also utilized the 3D structural information to improve the cross classifying specificities among the kinase-specific groups containing similar substrate motifs. |
The system flow of PhosK3D
| System Flow | 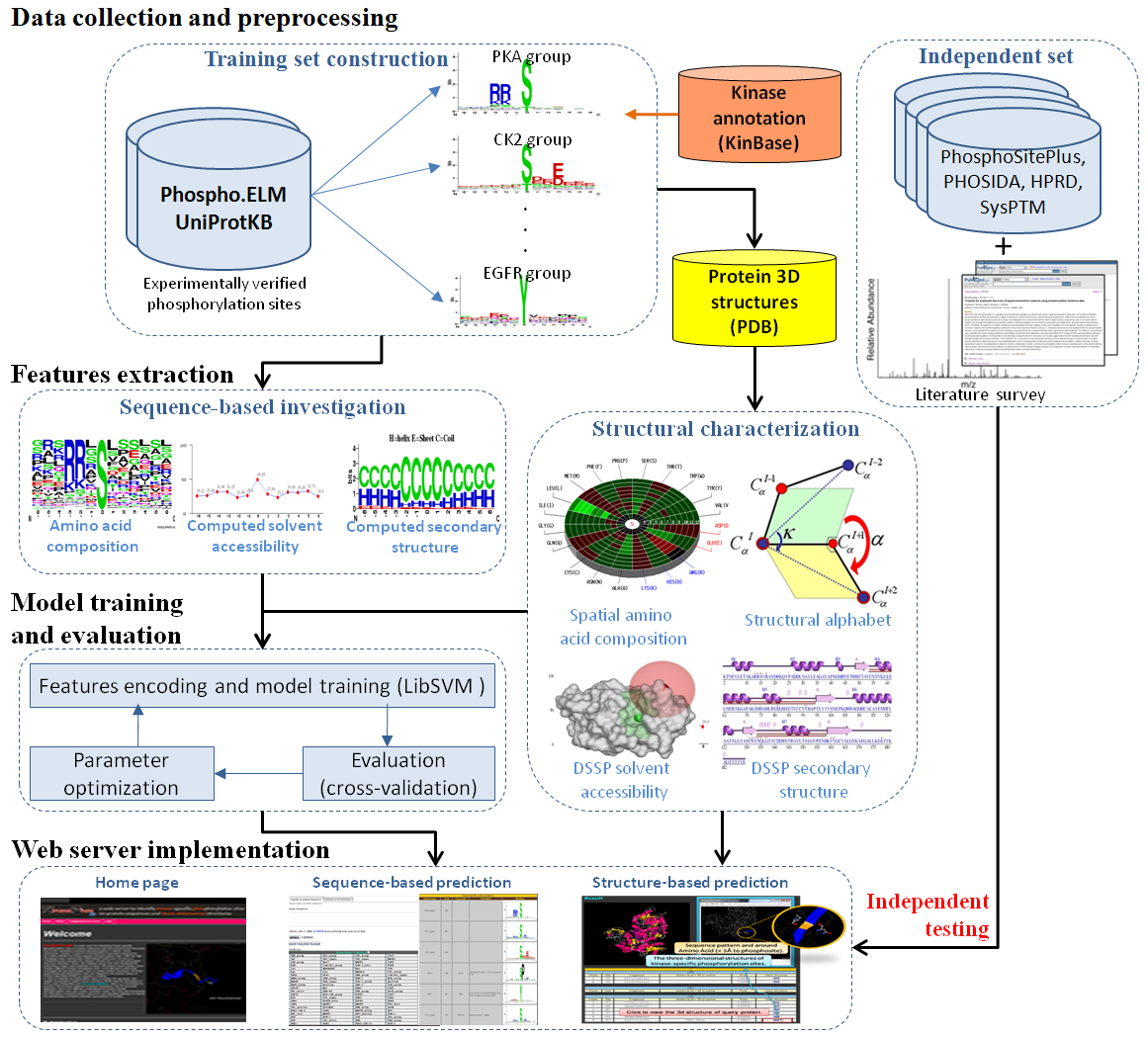 |