The highlighted on ubiquitin, the ubiquitin-proteasome Pathway, and crucial roles of E3 ligase:
♣ |
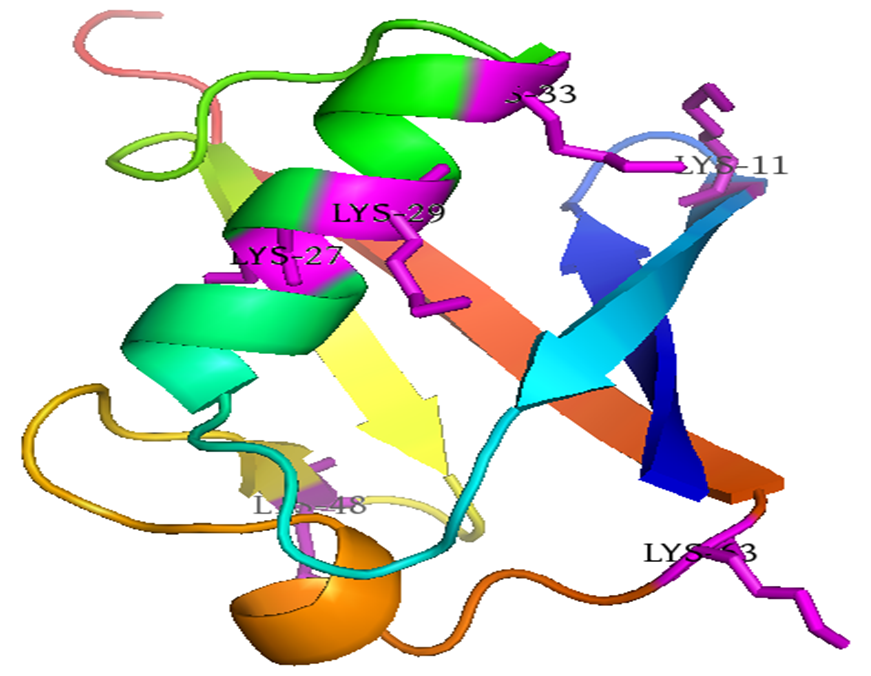
Ubiquitin is a small protein that consists of 76 amino acids with the weight of 8.5 kDa. Ubiquitin maintains its seven Lysine residues (Lys6, Lys11, Lys27, Lys29, Lys33, Lys48, and Lys63). The polyubiquitin can be formed through a bond between the C-terminal glycine of one ubiquitin molecule and an ϵ-amine of any one of the seven Lysine residues from another ubiquitin. |
| ♣ |
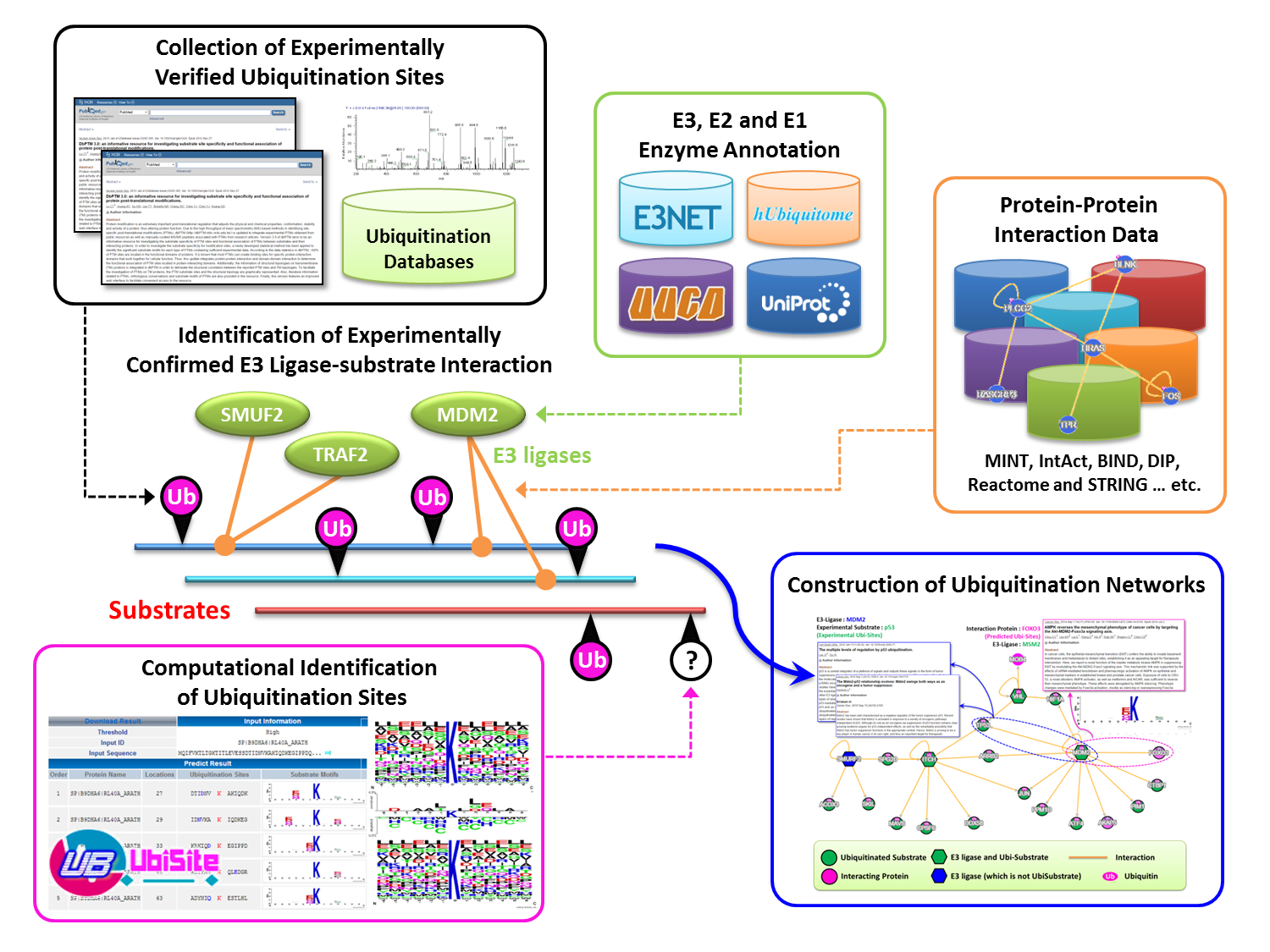
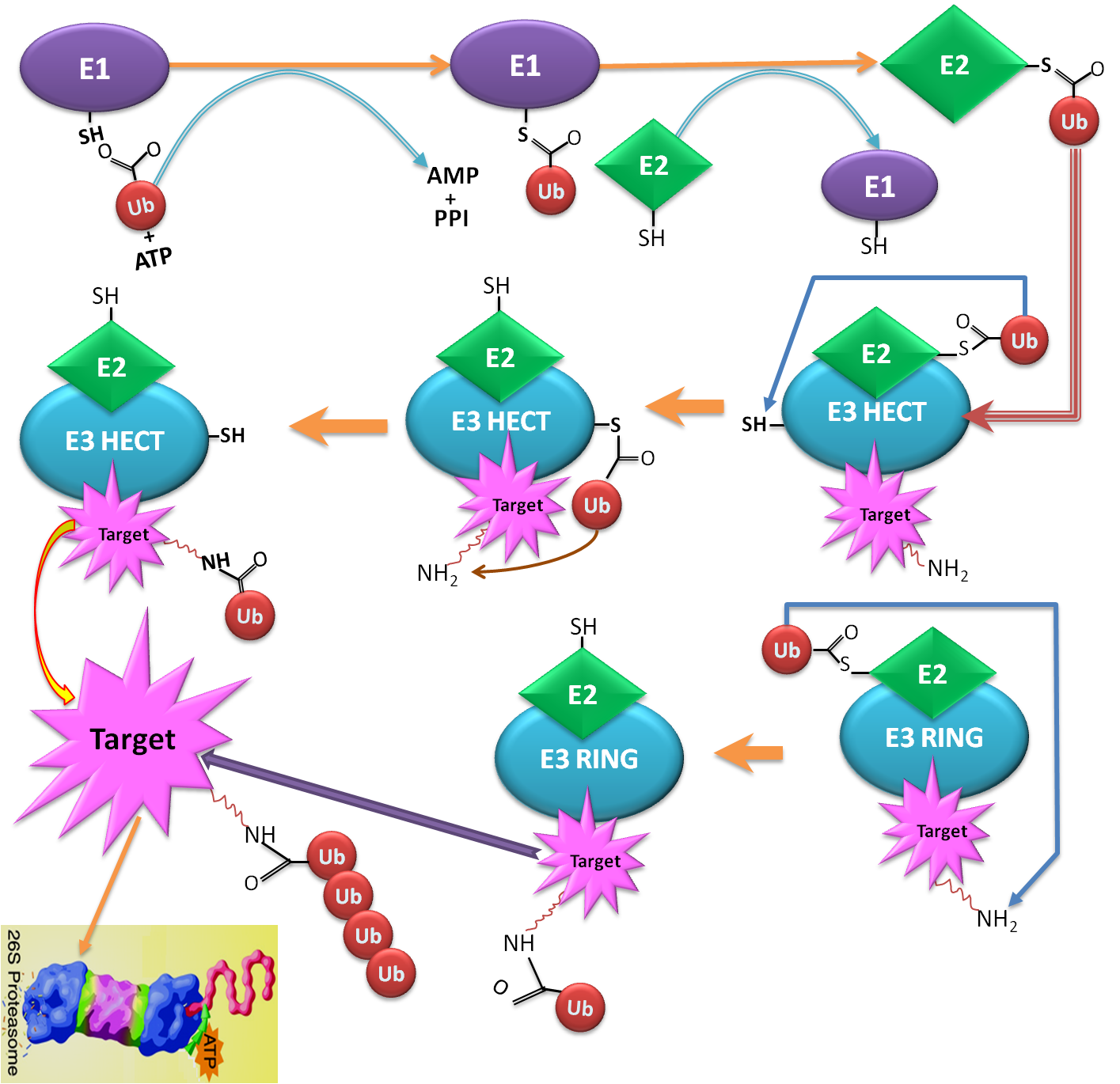
Ubiquitin conjugation sites of protein (also known as Ubiquitination or Ubiquitylation), an essential post-translational modification, is a sequential process that involves in three major enzymes including: E1 (activating enzyme), E2 (conjugating enzyme) and E3 (ubiquitin ligase). The ubiquitin-Proteasome Pathway is a complex and multi-step process, relating to a highly organized cascade of enzymatic reactions that select, mark, and degrades proteins. Firstly, the ubiquitination pathway is activated by attaching C-terminal residue of ubiquitin to a Cys sulphydryl residue in E1 enzyme. Secondly, ubiquitin attached to an E1 is transferred to an E2, which can be conjugated with various E3s. Ubiquitin-protein ligase E3 recognizes a specific protein substrate and catalyzes the transfer of activated ubiquitin to it. Finally, the substrate is sent to 26S proteasome for degradation |
| ♣ |
The ubiquitin ligase (E3 ligases) play a key role in regulating specific functions by recognizing or regulating a specific group of substrate proteins for ubiquitination. The presence of various different E3s and their substrate specificity indicate that the degradation process, which is one of the important regulatory mechanisms in cellular processes, is specifically controlled by E3s |
|
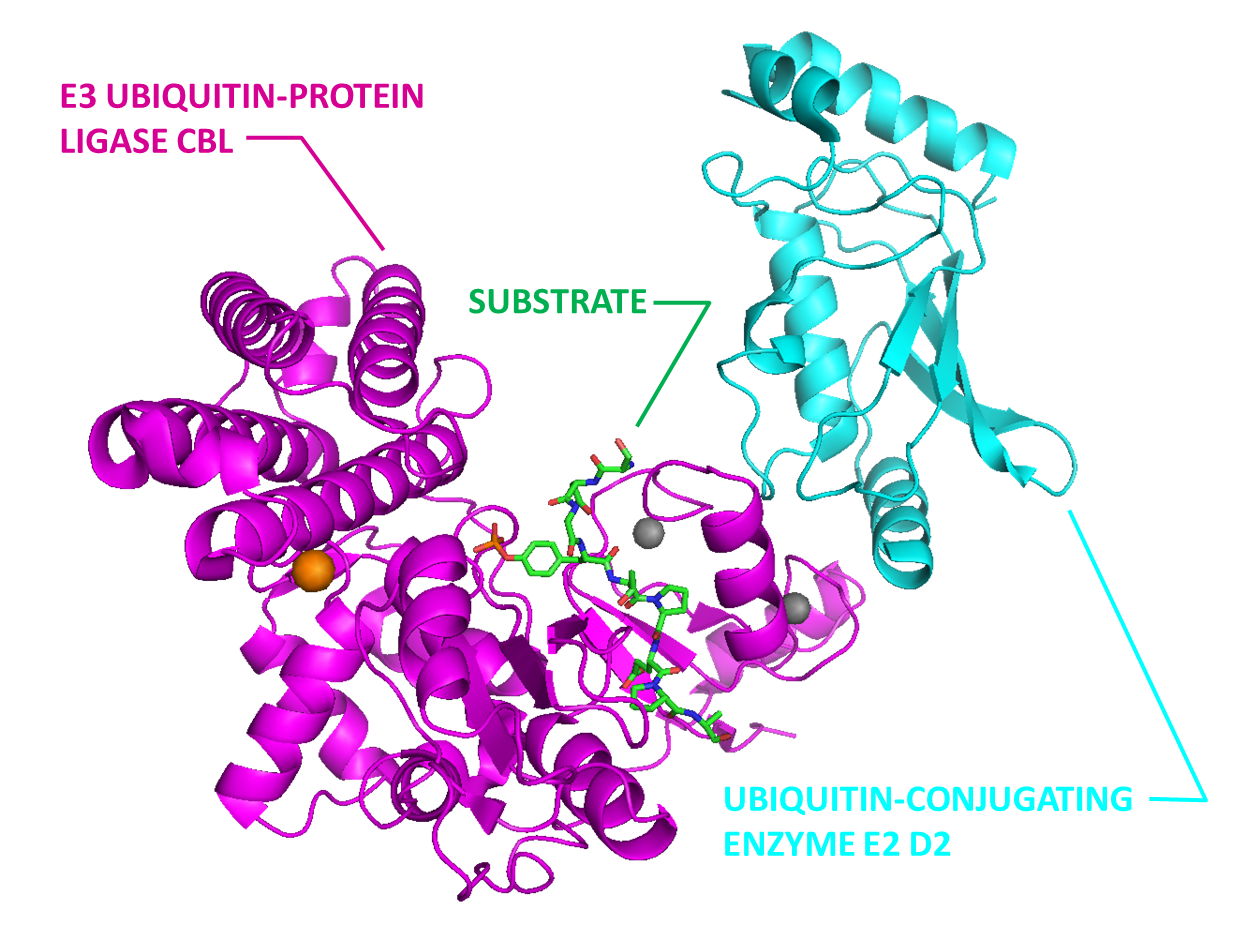 E3 ubiquitin ligase Cbl (Magentas) in complex with E2 (Cyan) and substrate peptide (Green).
E3 ubiquitin ligase Cbl (Magentas) in complex with E2 (Cyan) and substrate peptide (Green).