Protein Name: Probable E3 ubiquitin-protein ligase MYCBP2
|
UniprotKB/SwissProt ID: MYCB2_HUMAN (O75592)
Gene Name: MYCBP2
Synonyms: KIAA0916, PAM
Organism: Homo sapiens (Human).
Function: Probable E3 ubiquitin-protein ligase which mediates ubiquitination and subsequent proteasomal degradation of target proteins. May function as a facilitator or regulator of transcriptional activation by MYC. May have a role during synaptogenesis.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Nucleus.
| |
|
Graphical Visualization of Ubiquitination Sites:
|
| Overview of Protein Ubiquitination Sites with Functional and Structural Information | 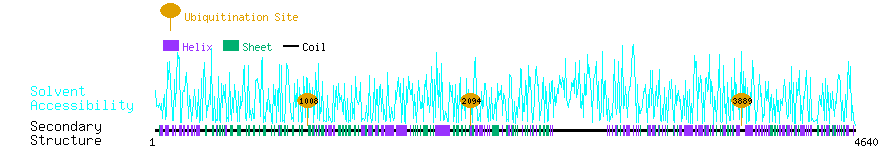 |
|

