Protein Name: E3 ubiquitin-protein ligase RNF169
|
UniprotKB/SwissProt ID: RN169_HUMAN (Q8NCN4)
Gene Name: RNF169
Synonyms: KIAA1991
Organism: Homo sapiens (Human).
Function: Probable E3 ubiquitin-protein ligase that acts as a negative regulator of double-strand breaks (DSBs) repair following DNA damage. Recruited to DSB repair sites by recognizing and binding ubiquitin catalyzed by RNF168 and competes with TP53BP1 and BRCA1 for association with RNF168-modified chromatin, thereby acting as a negative regulator of DSBs repair. E3 ubiquitin- protein ligase activity is not required for regulation of DSBs repair.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Nucleus, nucleoplasm. Note=Localizes to sites of double-strand breaks (DSBs) following DNA damage. Recruited to DSBs via recognition of RNF168-dependent ubiquitin products.
| |
|
Graphical Visualization of Ubiquitination Sites:
|
| Overview of Protein Ubiquitination Sites with Functional and Structural Information | 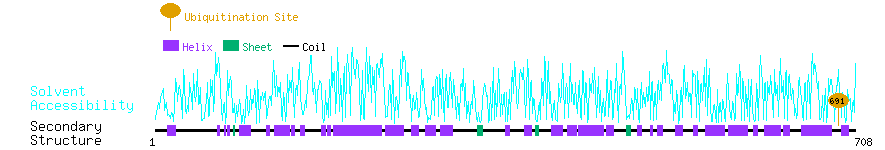 |
|

