Protein Name: Long-chain-fatty-acid--CoA ligase 3
|
UniprotKB/SwissProt ID: ACSL3_HUMAN (O95573)
Gene Name: ACSL3
Synonyms: ACS3, FACL3, LACS3
Organism: Homo sapiens (Human).
Function: Acyl-CoA synthetases (ACSL) activates long-chain fatty acids for both synthesis of cellular lipids, and degradation via beta-oxidation. ACSL3 mediates hepatic lipogenesis (By similarity). Preferentially uses myristate, laurate, arachidonate and eicosapentaenoate as substrates (By similarity). Has mainly an anabolic role in energy metabolism. Required for the incorporation of fatty acids into phosphatidylcholine, the major phospholipid located on the surface of VLDL (very low density lipoproteins).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Mitochondrion outer membrane; Single-pass type III membrane protein (By similarity). Peroxisome membrane; Single-pass type III membrane protein (By similarity). Microsome membrane; Single-pass type III membrane protein (By similarity). Endoplasmic reticu
| |
|
Protein disease:
|
| Disease database |
Database Entry |
Disease information | | OMIM | 602371 | ACYL-CoA SYNTHETASE LONG-CHAIN FAMILY MEMBER 3; ACSL3 ;;FATTY ACID CoA LIGASE, LONG-CHAIN 3; |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 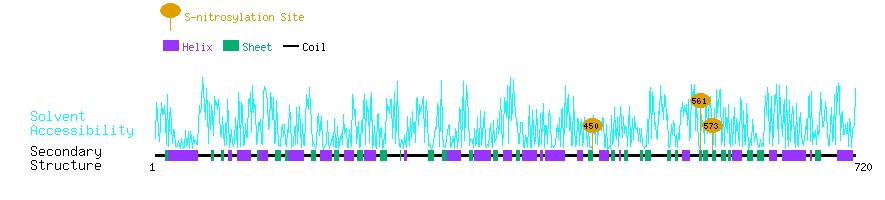 |
|

