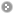|
How to Link:
Users can directly link to dbSNO by SwissProt ID.
For example:
PTM Resources:
- UniProt KB
- PhosphoSite
- dbPTM 3.0
- dbGSH
- RegPhos 2.0
- KinasePhos
- Phospho.ELM
- OGlycBase
Version: 2.0
(Sep. 15, 2014)
|
|
|
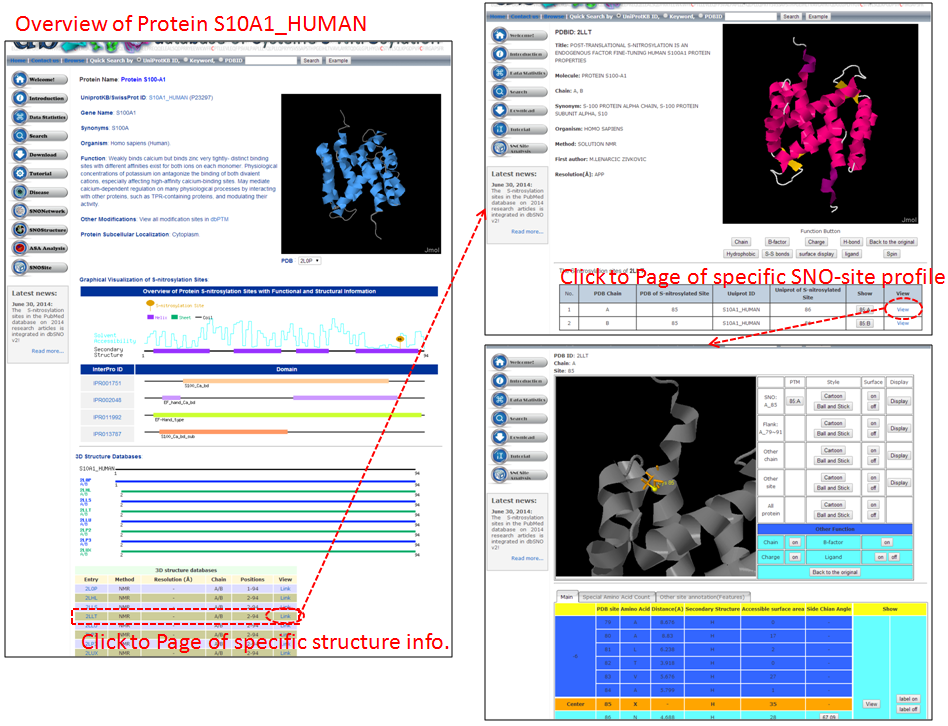
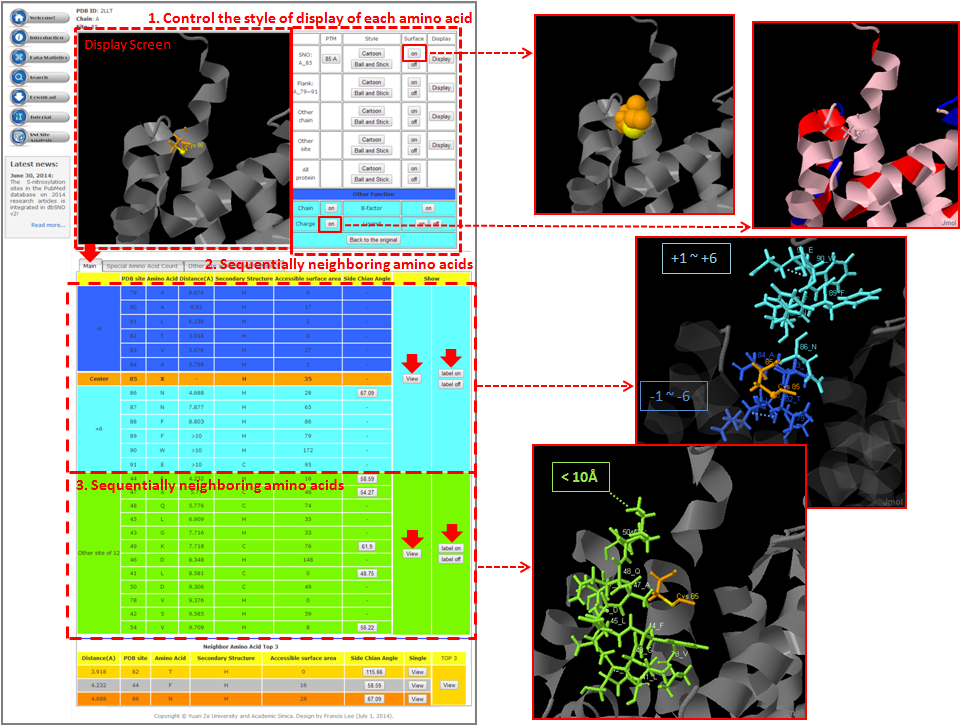
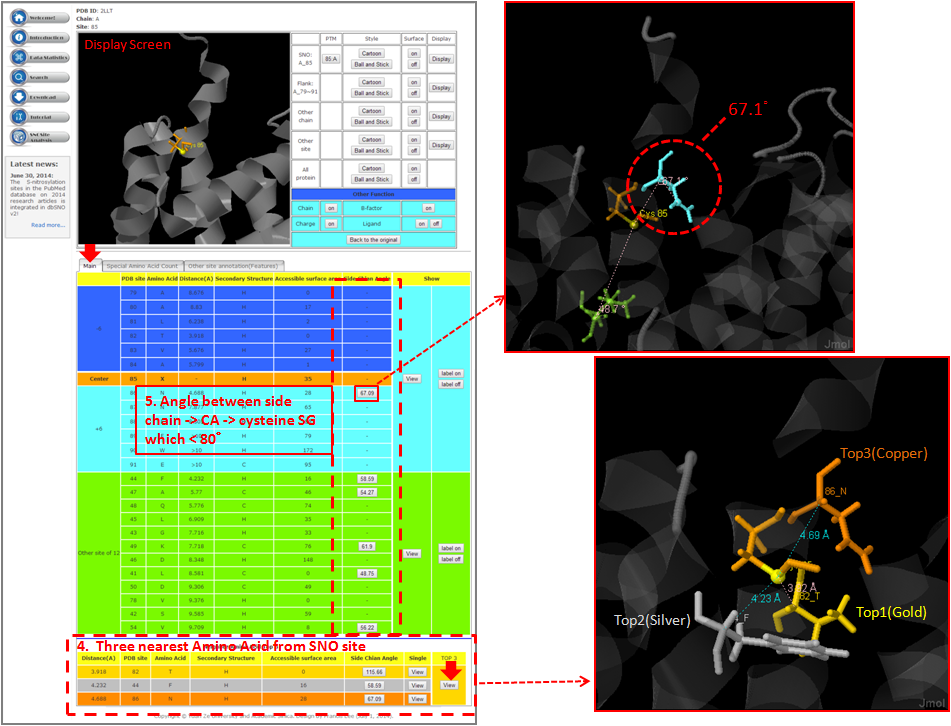
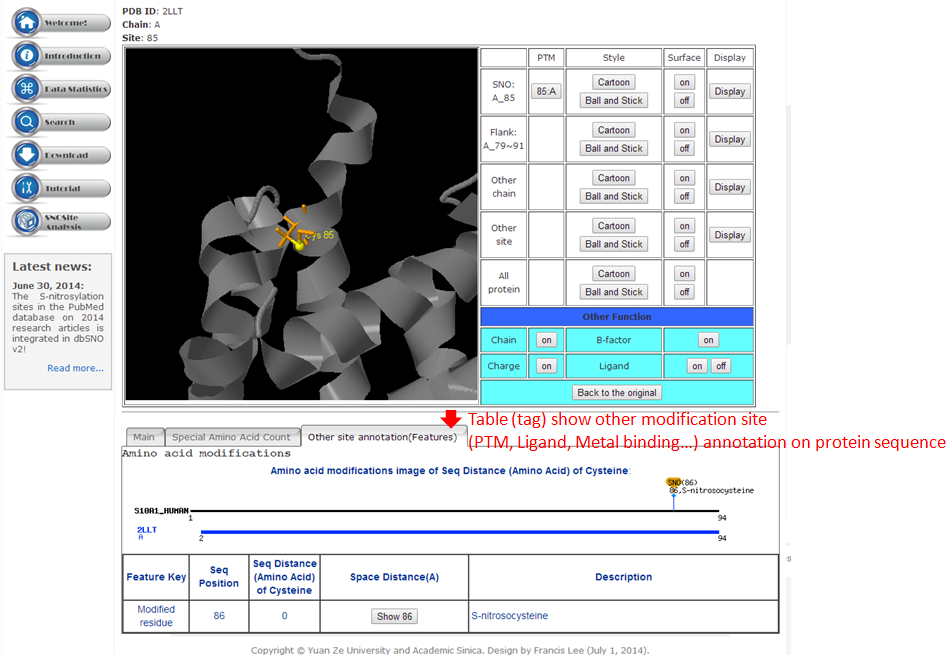
Step 1. Three major queries in search page. Users can query their interested protein by input the protein name, gene name, UniProtKB ID, AC, PDB ID, InterPro ID in search field of homepage. It was also allowed the users to query the amino acid sequence against the release 2014_07 of Swiss-Prot protein sequences. 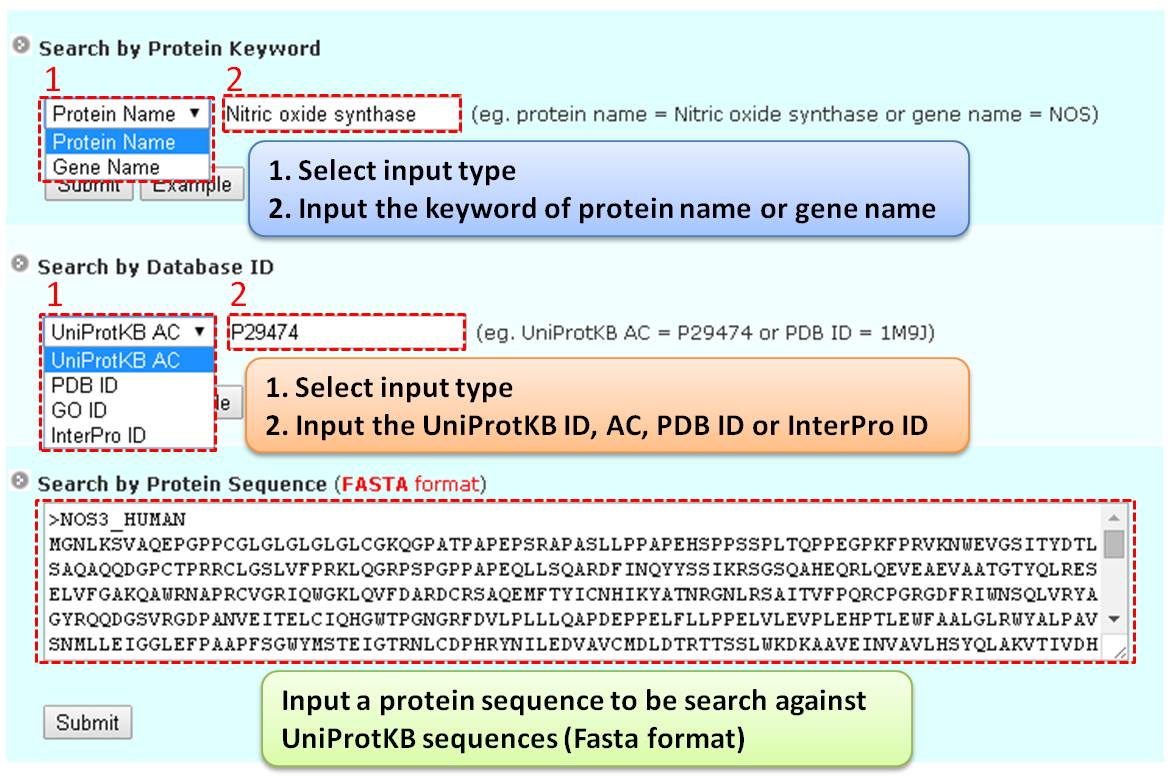 Step 2. The search result. Since users input the keyword of protein name or gene name, UniProtKB ID, AC, PDB ID, InterPro ID in search field of homepage. The proteins matching the query keywords were shown in a table. Then, users can click on the URL link "show" to view the detailed information about the protein, such as protein function, S-nitrosylation sites, protein doamin, etc. 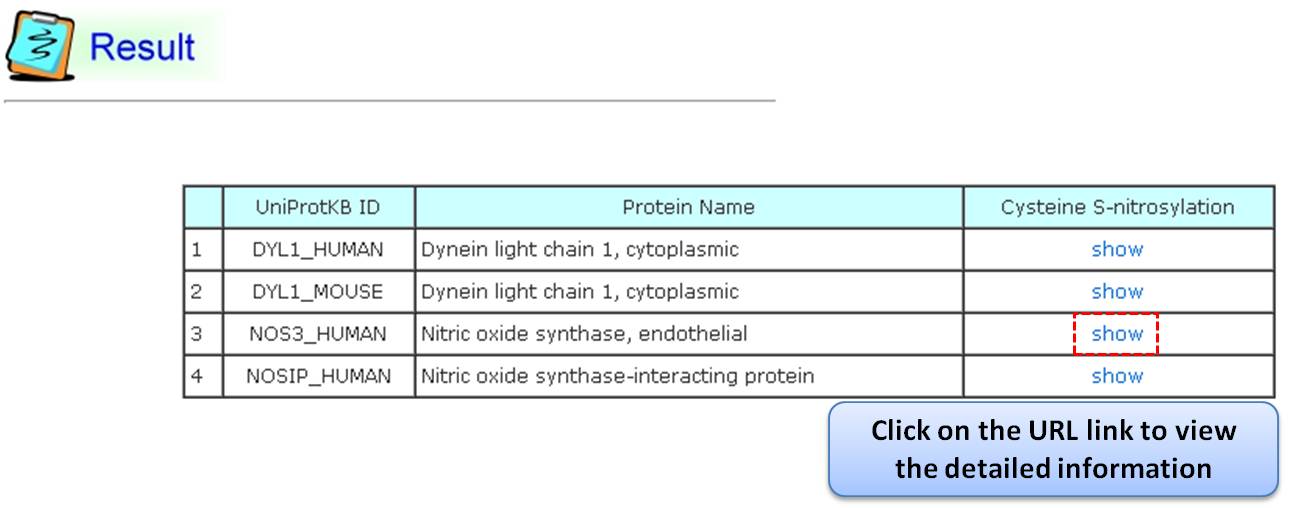 Step 3. The detail information about S-nitrosylation. Users can view the experimental S-nitrosylation sites and graphical visualizations. Graphical visualization could reveal an overview of the S-nitrosylation sites, the solvent accessibility of the residues, protein variations, protein secondary structures and protein functional domains in protein sequence. (a) Quick search by IDs and keywords. (b) Basic information. (c) Visualization of tertiary structure. (d) Protein disease information. (e) Network with metabolic pathway. (f) Graphical visualization of S-nitrosylation sites with structural characteristics and functional domains. (g) NO Site Structure Analysis. (h) Table of S-nitrosylation sites with reported literatures.  Step 4. The substrate motifs conserved regions. Users can investigate whether a S-nitrosylation site located in substrate motifs conserved regions by click on the URL in the table of S-nitrosylation sites. 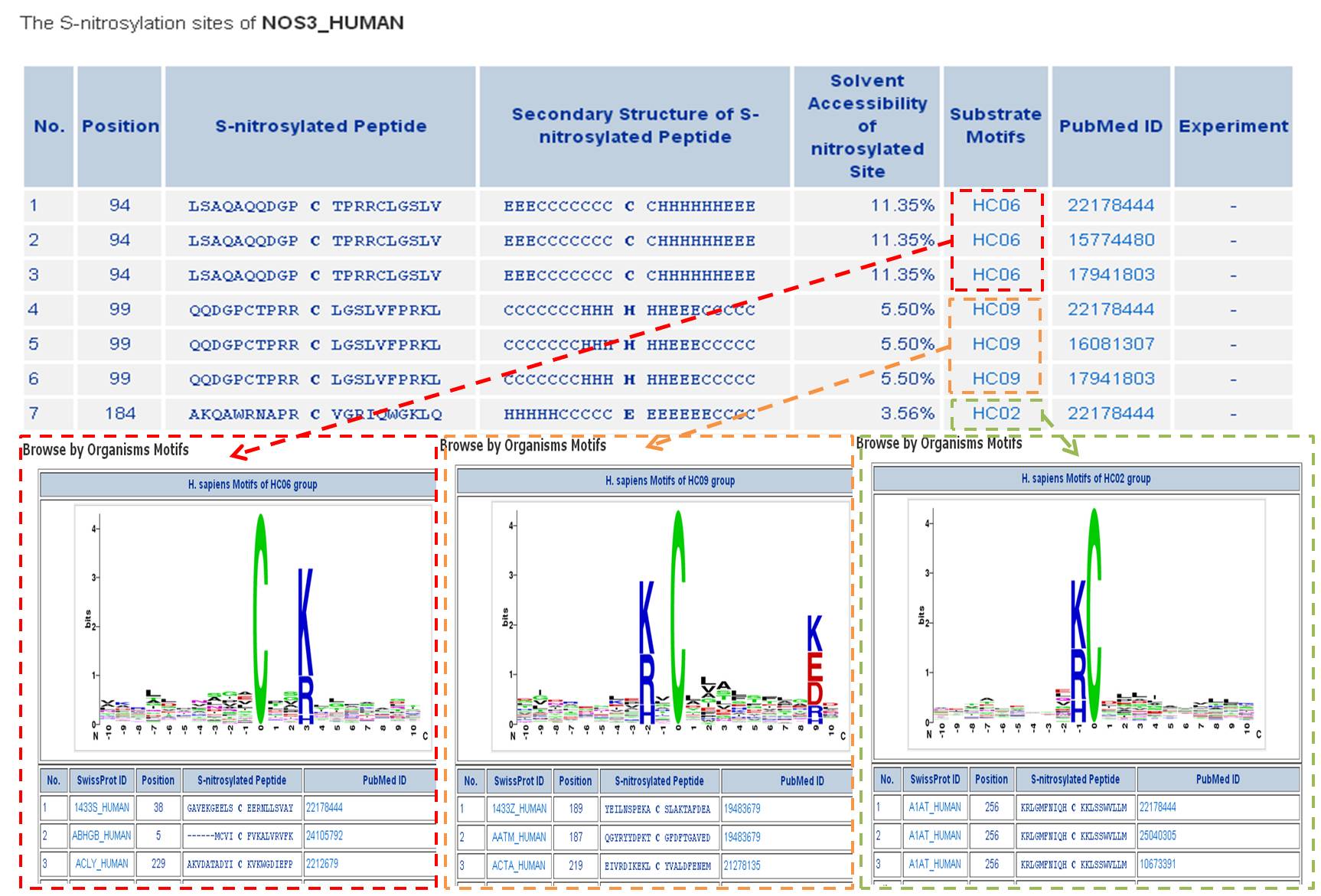
Step 1. The summary table of S-nitrosylation. To facilitate the investigation in each type of S-nitrosylation, we developed a summary table for users to browse all the types of S-nitrosylation in the release 2.0 of dbSNO. In the table, each type of S-nitrosylation was categorized by their modified amino acids with the number of experimentally verified sites. For example, users can choose the M. musculus (Mouse) to take the more detailed information such as the position of modification on amino acid, the location of modification on protein sequence, the modified chemical formula, and the mass difference. 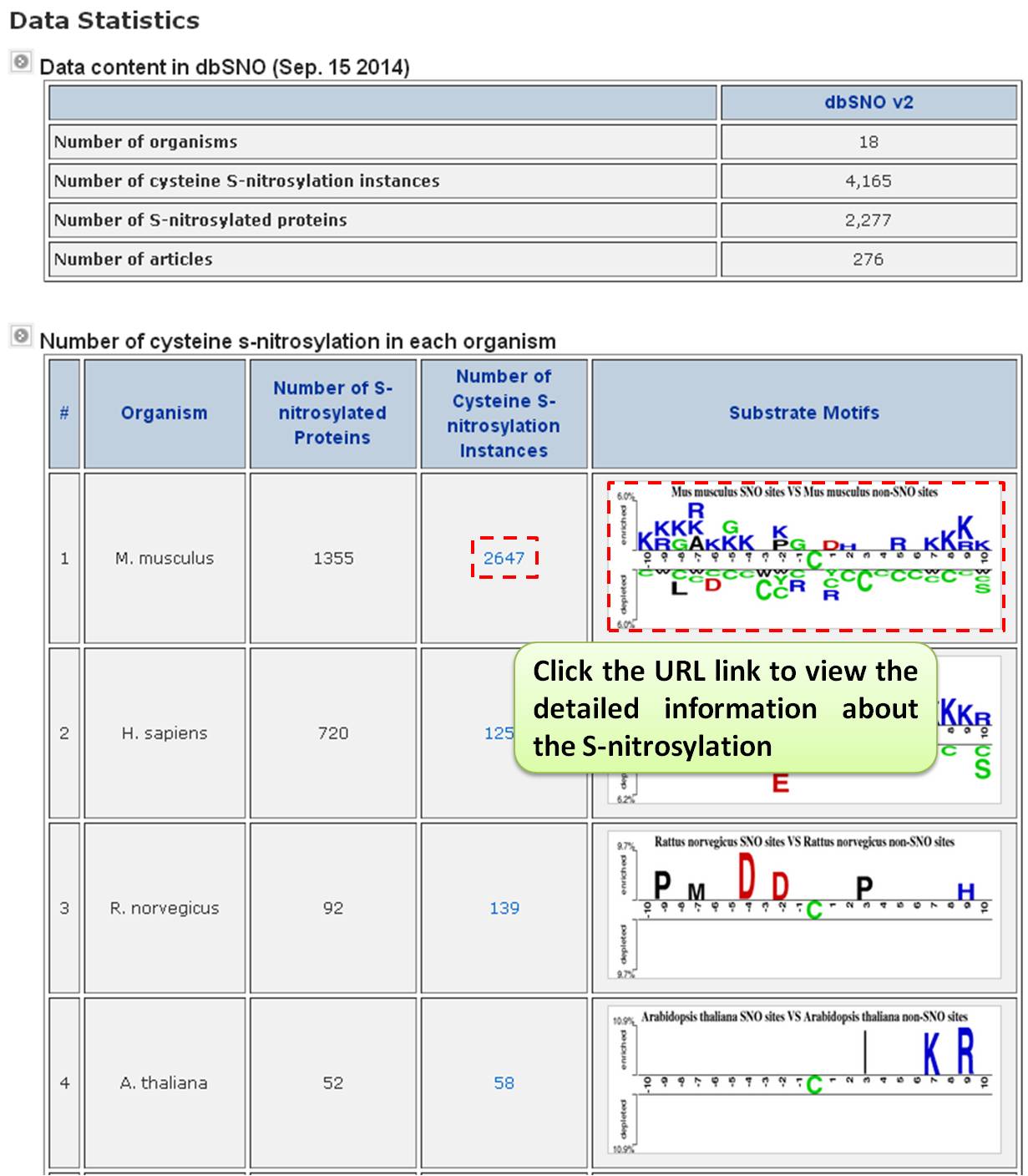 Step 2. Users can investigate the detailed information M. musculus (Mouse) on proteins, such as the position of modification on amino acid, the location of modification on protein sequence, the modified chemical formula, and the mass difference. Especially, the formula structure was visualized by the Jmol program which was implemented in JAVA Applet. However, the most effective knowledge about the S-nitrosylation is the substrate site specificity including the frequency of amino acids, the average solvent accessibility, and the frequency of secondary structure surrounding the modified site. The sequence logo which represents the frequency of amino acids and the frequency of secondary structure surrounding the modified sites was provided. 
This Page, we used structure alignment method to compare the difference between N=O binding and N=O Non-binding yet in same protein and on same SNO site, this purpose effect of N=O binding in protein structure. 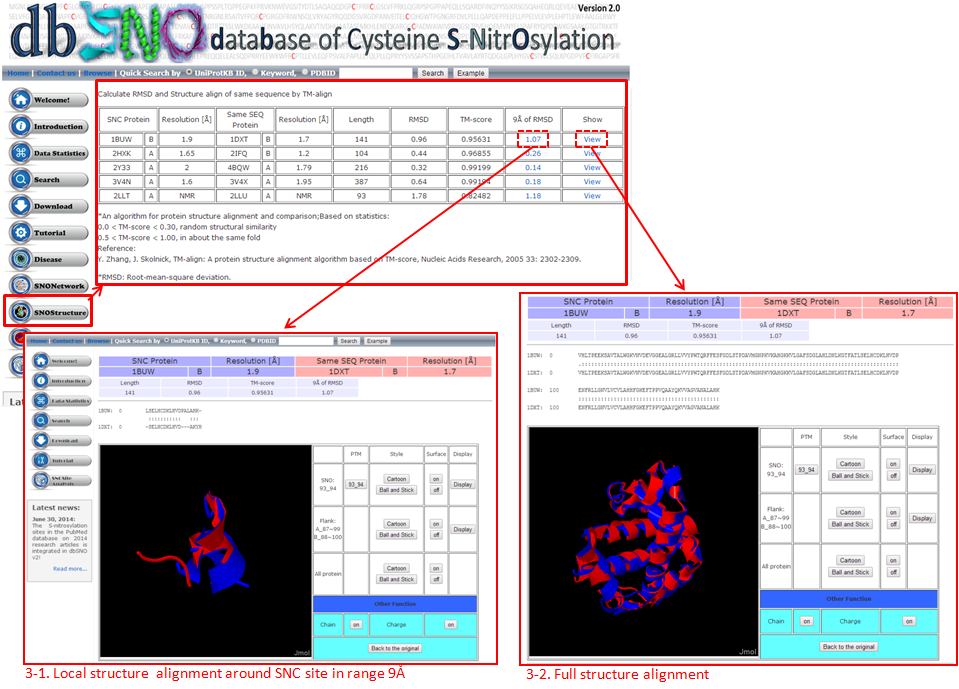 3-1. Local structure alignment around SNC site in range 9Å. 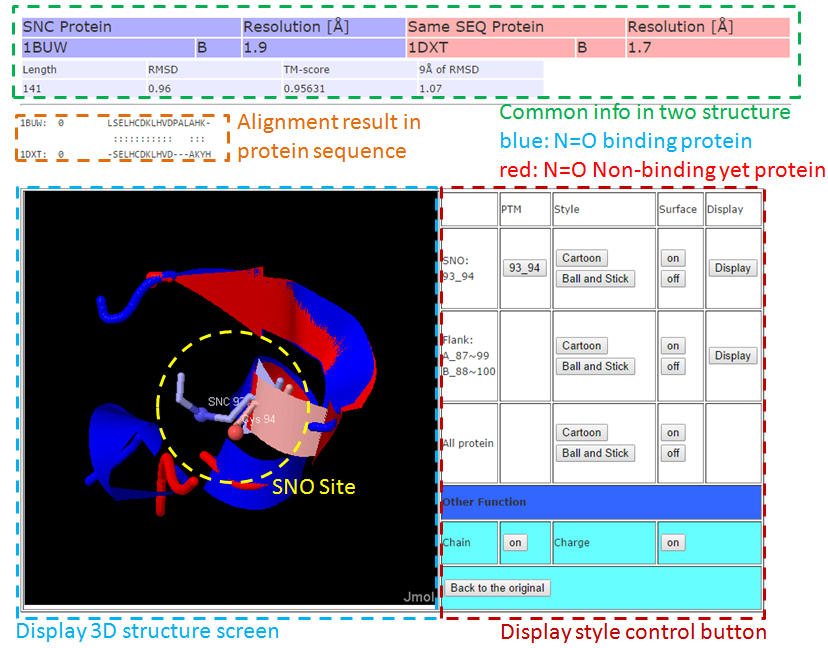 3-2. Full structure alignment. 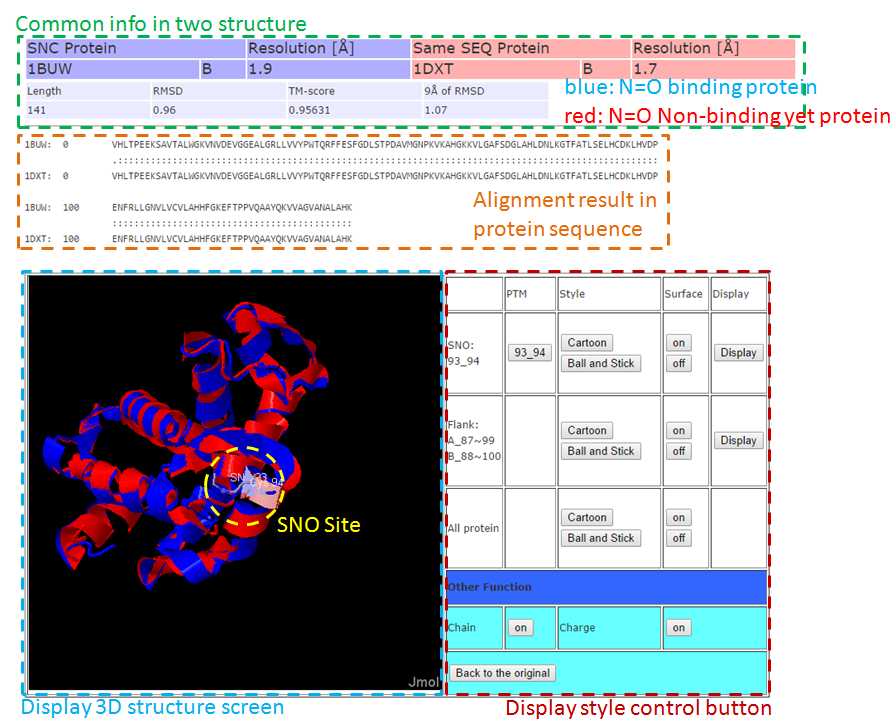
    |