Protein Name: Acyl-coenzyme A synthetase ACSM1, mitochondrial
|
UniprotKB/SwissProt ID: ACSM1_MOUSE (Q91VA0)
Gene Name: Acsm1
Synonyms: Bucs1, Lae, Macs1
Organism: Mus musculus (Mouse).
Function: Functions as GTP-dependent lipoate-activating enzyme that generates the substrate for lipoyltransferase (By similarity). Has medium-chain fatty acid:CoA ligase activity with broad substrate specificity (in vitro). Acts on acids from C(4) to C(11) and on the corresponding 3-hydroxy- and 2,3- or 3,4- unsaturated acids (in vitro).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Mitochondrion matrix.
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 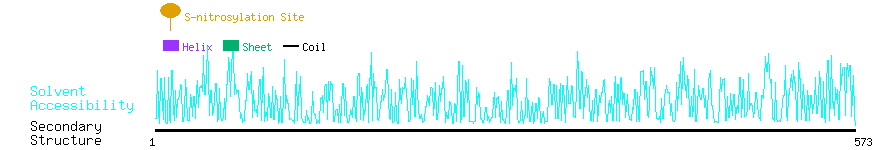 |
|

