Protein Name: Sarcoplasmic/endoplasmic reticulum calcium ATPase 2
|
UniprotKB/SwissProt ID: AT2A2_MOUSE (O55143)
Gene Name: Atp2a2
Organism: Mus musculus (Mouse).
Function: This magnesium-dependent enzyme catalyzes the hydrolysis of ATP coupled with the translocation of calcium from the cytosol to the sarcoplasmic reticulum lumen. Isoform SERCA2A is involved in the regulation of the contraction/relaxation cycle (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Endoplasmic reticulum membrane; Multi-pass membrane protein. Sarcoplasmic reticulum membrane; Multi-pass membrane protein.
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 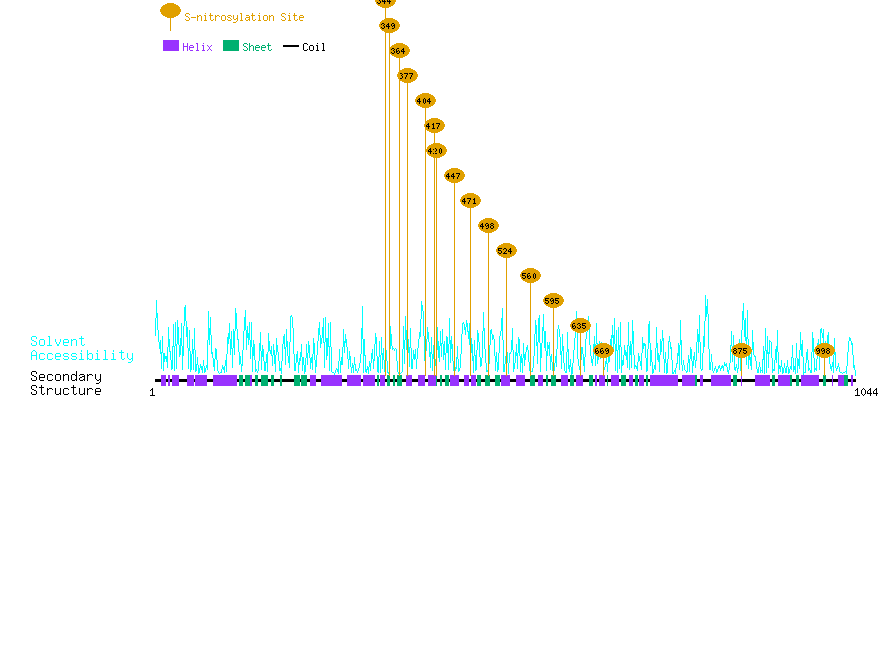 |
|

