Protein Name: Charged multivesicular body protein 5
|
UniprotKB/SwissProt ID: CHMP5_MOUSE (Q9D7S9)
Gene Name: Chmp5
Synonyms: Snf7dc2
Organism: Mus musculus (Mouse).
Function: Probable peripherally associated component of the endosomal sorting required for transport complex III (ESCRT-III) which is involved in multivesicular bodies (MVBs) formation and sorting of endosomal cargo proteins into MVBs. MVBs contain intraluminal vesicles (ILVs) that are generated by invagination and scission from the limiting membrane of the endosome and mostly are delivered to lysosomes enabling degradation of membrane proteins, such as stimulated growth factor receptors, lysosomal enzymes and lipids. The MVB pathway appears to require the sequential function of ESCRT-O, -I,-II and -III complexes. ESCRT- III proteins mostly dissociate from the invaginating membrane before the ILV is released. The ESCRT machinery also functions in topologically equivalent membrane fission events, such as the terminal stages of cytokinesis. ESCRT-III proteins are believed to mediate the necessary vesicle extrusion and/or membrane fission activities, possibly in conjunction with the AAA ATPase VPS4 (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cytoplasm, cytosol (By similarity). Endosome membrane; Peripheral membrane protein (By similarity). Note=Localizes to the midbody of dividing cells. Localized in two distinct rings on either side of the Fleming body (By similarity).
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 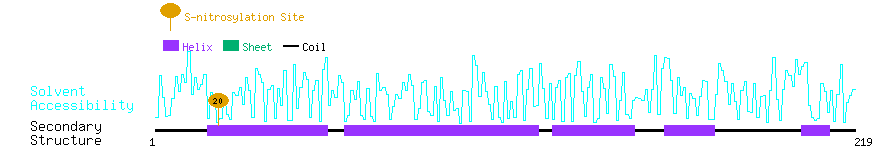 |
|

