Protein Name: Catenin alpha-1
|
UniprotKB/SwissProt ID: CTNA1_HUMAN (P35221)
Gene Name: CTNNA1
Organism: Homo sapiens (Human).
Function: Associates with the cytoplasmic domain of a variety of cadherins. The association of catenins to cadherins produces a complex which is linked to the actin filament network, and which seems to be of primary importance for cadherins cell-adhesion properties. Can associate with both E- and N-cadherins. Originally believed to be a stable component of E-cadherin/catenin adhesion complexes and to mediate the linkage of cadherins to the actin cytoskeleton at adherens junctions. In contrast, cortical actin was found to be much more dynamic than E-cadherin/catenin complexes and CTNNA1 was shown not to bind to F-actin when assembled in the complex suggesting a different linkage between actin and adherens junctions components. The homodimeric form may regulate actin filament assembly and inhibit actin branching by competing with the Arp2/3 complex for binding to actin filaments. May play a crucial role in cell differentiation.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Isoform 1: Cytoplasm, cytoskeleton. Cell junction, adherens junction. Cell membrane; Peripheral membrane protein; Cytoplasmic side. Cell junction. Note=Found at cell-cell boundaries and probably at cell-matrix boundaries. Isoform 3: Cell membrane; Periph
|
PDB :
( If your security settings prevent Jmol from running, please register http://140.138.144.145/ as a safe location in your Java settings. ) |
|
Protein disease:
|
| Disease database |
Database Entry |
Disease information | | OMIM | 116805 | CATENIN, ALPHA-1; CTNNA1 ;;ALPHA-E-CATENIN;; CADHERIN-ASSOCIATED PROTEIN |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 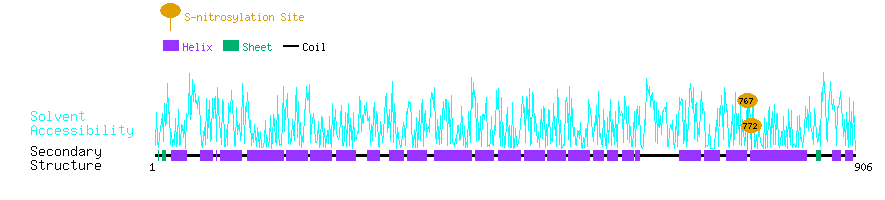 |
|

