Protein Name: Cytochrome c, somatic
|
UniprotKB/SwissProt ID: CYC_MOUSE (P62897)
Gene Name: Cycs
Organism: Mus musculus (Mouse).
Function: Electron carrier protein. The oxidized form of the cytochrome c heme group can accept an electron from the heme group of the cytochrome c1 subunit of cytochrome reductase. Cytochrome c then transfers this electron to the cytochrome oxidase complex, the final protein carrier in the mitochondrial electron-transport chain. Plays a role in apoptosis. Suppression of the anti- apoptotic members or activation of the pro-apoptotic members of the Bcl-2 family leads to altered mitochondrial membrane permeability resulting in release of cytochrome c into the cytosol. Binding of cytochrome c to Apaf-1 triggers the activation of caspase-9, which then accelerates apoptosis by activating other caspases.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Mitochondrion intermembrane space. Note=Loosely associated with the inner membrane.
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 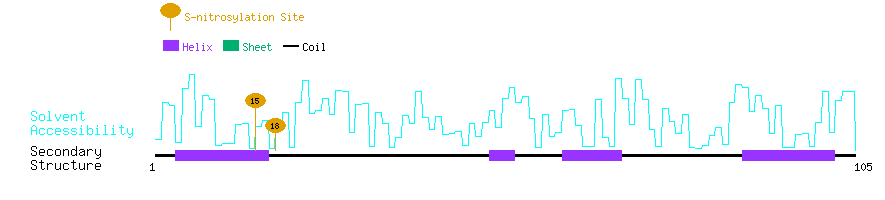 |
|

