Protein Name: Malonyl-CoA decarboxylase, mitochondrial
|
UniprotKB/SwissProt ID: DCMC_MOUSE (Q99J39)
Gene Name: Mlycd
Organism: Mus musculus (Mouse).
Function: Catalyzes the conversion of malonyl-CoA to acetyl-CoA. In the fatty acid biosynthesis MCD selectively removes malonyl-CoA and thus assures that methyl-malonyl-CoA is the only chain elongating substrate for fatty acid synthase and that fatty acids with multiple methyl side chains are produced. In peroxisomes it may be involved in degrading intraperoxisomal malonyl-CoA, which is generated by the peroxisomal beta-oxidation of odd chain-length dicarboxylic fatty acids. Plays a role in the metabolic balance between glucose and lipid oxidation in muscle independent of alterations in insulin signaling. Plays a role in controlling the extent of ischemic injury by promoting glucose oxidation.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cytoplasm (By similarity). Mitochondrion matrix. Peroxisome (By similarity). Peroxisome matrix (By similarity). Note=Enzymatically active in all three subcellular compartments (By similarity).
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 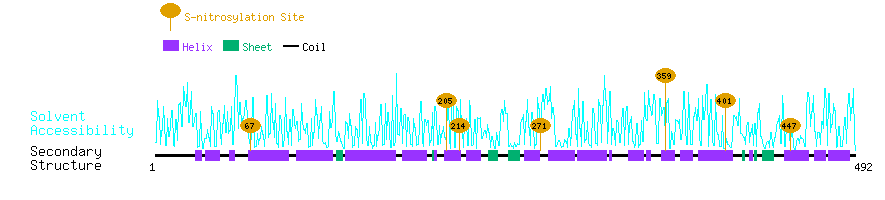 |
|

