Protein Name: Dehydrogenase/reductase SDR family member 4
|
UniprotKB/SwissProt ID: DHRS4_MOUSE (Q99LB2)
Gene Name: Dhrs4
Synonyms: D14Ucla2
Organism: Mus musculus (Mouse).
Function: Reduces all-trans-retinal and 9-cis retinal. Can also catalyze the oxidation of all-trans-retinol with NADP as co- factor, but with much lower efficiency. Reduces alkyl phenyl ketones and alpha-dicarbonyl compounds with aromatic rings, such as pyrimidine-4-aldehyde, 3-benzoylpyridine, 4-benzoylpyridine, menadione and 4-hexanoylpyridine. Has no activity towards aliphatic aldehydes and ketones (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Peroxisome (By similarity).
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 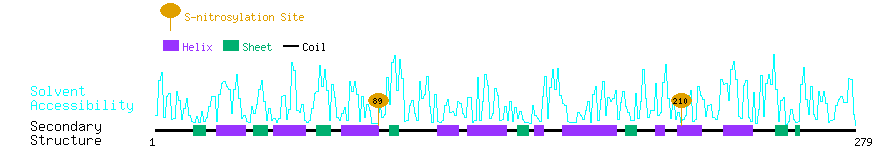 |
|

