Protein Name: Deleted in malignant brain tumors 1 protein
|
UniprotKB/SwissProt ID: DMBT1_HUMAN (Q9UGM3)
Gene Name: DMBT1
Synonyms: GP340
Organism: Homo sapiens (Human).
Function: May be considered as a candidate tumor suppressor gene for brain, lung, esophageal, gastric, and colorectal cancers. May play roles in mucosal defense system, cellular immune defense and epithelial differentiation. May play a role as an opsonin receptor for SFTPD and SPAR in macrophage tissues throughout the body, including epithelial cells lining the gastrointestinal tract. May play a role in liver regeneration. May be an important factor in fate decision and differentiation of transit-amplifying ductular (oval) cells within the hepatic lineage. Required for terminal differentiation of columnar epithelial cells during early embryogenesis. May function as a binding protein in saliva for the regulation of taste sensation. Binds to HIV-1 envelope protein and has been shown to both inhibit and facilitate viral transmission. Displays a broad calcium-dependent binding spectrum against both Gram-positive and Gram-negative bacteria, suggesting a role in defense against bacterial pathogens. Binds to a range of poly- sulfated and poly-phosphorylated ligands which may explain its broad bacterial-binding specificity. Inhibits cytoinvasion of S.enterica. Associates with the actin cytoskeleton and is involved in its remodeling during regulated exocytosis. Interacts with pancreatic zymogens in a pH-dependent manner and may act as a Golgi cargo receptor in the regulated secretory pathway of the pancreatic acinar cell.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Secreted (By similarity). Note=Some isoforms may be membrane-bound. Localized to the lumenal aspect of crypt cells in the small intestine. In the colon, seen in the lumenal aspect of surface epithelial cells. Formed in the ducts of von Ebner gland, and r
| |
|
Protein disease:
|
| Disease database |
Database Entry |
Disease information | | HPRD | 03575 | Glioblastoma multiforme, somatic |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 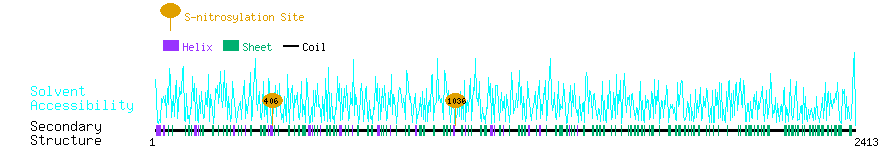 |
|

