Protein Name: Dynamin-1-like protein
|
UniprotKB/SwissProt ID: DNM1L_MOUSE (Q8K1M6)
Gene Name: Dnm1l
Synonyms: Drp1
Organism: Mus musculus (Mouse).
Function: Functions in mitochondrial and peroxisomal division. Mediates membrane fission through oligomerization into ring-like structures which wrap around the scission site to constict and sever the mitochondrial membrane through a GTP hydrolysis- dependent mechanism. Through its function in mitochondrial division, ensures the survival of at least some types of postmitotic neurons, including Purkinje cells, by suppressing oxidative damage. Required for normal brain development, including that of cerebellum. Facilitates developmentally regulated apoptosis during neural tube formation. Required for a normal rate of cytochrome c release and caspase activation during apoptosis; this requirement may depend upon the cell type and the physiological apoptotic cues. Also required for mitochondrial fission during mitosis. Required for formation of endocytic vesicles. Proposed to regulate synaptic vesicle membrane dynamics through association with BCL2L1 isoform Bcl-X(L) which stimulates its GTPase activity in synaptic vesicles; the function may require its recruitment by MFF to clathrin-containing vesicles. Required for programmed necrosis execution.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cytoplasm, cytosol. Golgi apparatus (By similarity). Endomembrane system; Peripheral membrane protein. Mitochondrion outer membrane; Peripheral membrane protein (By similarity). Peroxisome (By similarity). Membrane, clathrin-coated pit (By similarity). C
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 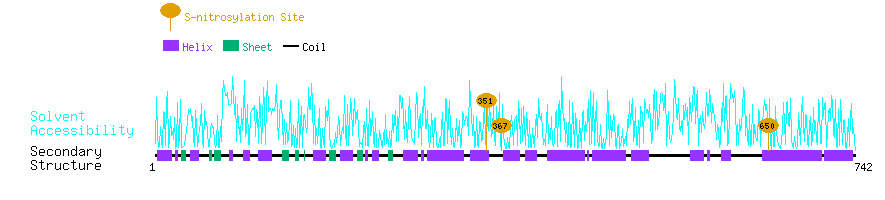 |
|

