Protein Name: Epidermal growth factor receptor
|
UniprotKB/SwissProt ID: EGFR_HUMAN (P00533)
Gene Name: EGFR
Synonyms: ERBB, ERBB1, HER1
Organism: Homo sapiens (Human).
Function: Receptor tyrosine kinase binding ligands of the EGF family and activating several signaling cascades to convert extracellular cues into appropriate cellular responses. Known ligands include EGF, TGFA/TGF-alpha, amphiregulin, epigen/EPGN, BTC/betacellulin, epiregulin/EREG and HBEGF/heparin-binding EGF. Ligand binding triggers receptor homo- and/or heterodimerization and autophosphorylation on key cytoplasmic residues. The phosphorylated receptor recruits adapter proteins like GRB2 which in turn activates complex downstream signaling cascades. Activates at least 4 major downstream signaling cascades including the RAS- RAF-MEK-ERK, PI3 kinase-AKT, PLCgamma-PKC and STATs modules. May also activate the NF-kappa-B signaling cascade. Also directly phosphorylates other proteins like RGS16, activating its GTPase activity and probably coupling the EGF receptor signaling to the G protein-coupled receptor signaling. Also phosphorylates MUC1 and increases its interaction with SRC and CTNNB1/beta-catenin. Isoform 2 may act as an antagonist of EGF action.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cell membrane; Single-pass type I membrane protein. Endoplasmic reticulum membrane; Single-pass type I membrane protein. Golgi apparatus membrane; Single-pass type I membrane protein. Nucleus membrane; Single-pass type I membrane protein. Endosome. Endos
|
PDB :
( If your security settings prevent Jmol from running, please register http://140.138.144.145/ as a safe location in your Java settings. ) |
|
Protein disease:
|
| Disease database |
Database Entry |
Disease information | | KEGG | H00016 | Oral cancer | | KEGG | H00017 | Esophageal cancer | | KEGG | H00018 | Gastric cancer | | KEGG | H00022 | Bladder cancer | | KEGG | H00028 | Choriocarcinoma | | KEGG | H00030 | Cervical cancer | | KEGG | H00042 | Glioma | | KEGG | H00055 | Laryngeal cancer | | OMIM | 131550 | EPIDERMAL GROWTH FACTOR RECEPTOR; EGFR ;;V-ERB-B AVIAN ERYTHROBLASTIC LEUKEMIA VIRAL ONCOGEN | | OMIM | 211980 | LUNG CANCER ALVEOLAR CELL CARCINOMA, INCLUDED;; ADENOCARCINOMA OF LUNG, INCLUDED;; NONSMALL | | HPRD | 00579 | Nonsmall cell lung cancer, resistance to tyrosine kinase inhibitor in | | HPRD | 00579 | Nonsmall cell lung cancer, response to tyrosine kinase inhibitor in, somatic |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 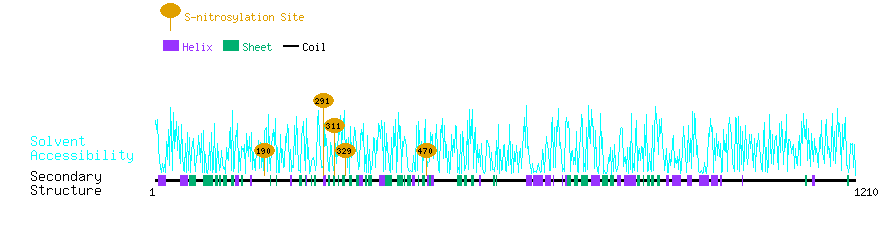 |
|
