Protein Name: Alpha-enolase
|
UniprotKB/SwissProt ID: ENOA_MOUSE (P17182)
Gene Name: Eno1
Synonyms: Eno-1
Organism: Mus musculus (Mouse).
Function: Multifunctional enzyme that, as well as its role in glycolysis, plays a part in various processes such as growth control, hypoxia tolerance and allergic responses (By similarity). May also function in the intravascular and pericellular fibrinolytic system due to its ability to serve as a receptor and activator of plasminogen on the cell surface of several cell-types such as leukocytes and neurons. Stimulates immunoglobulin production (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cytoplasm (By similarity). Cell membrane (By similarity). Note=Can translocate to the plasma membrane in either the homodimeric (alpha/alpha) or heterodimeric (alpha/gamma) form (By similarity). ENO1 is localized to the M-band.
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 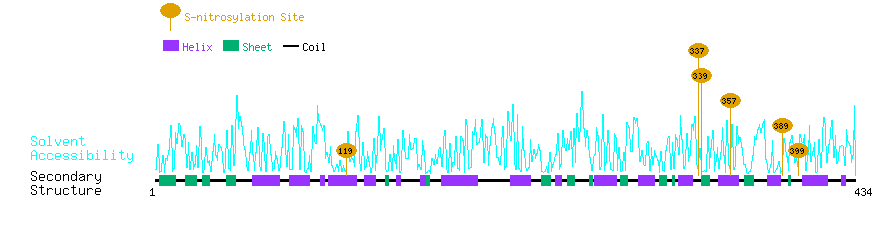 |
|

