Protein Name: Eukaryotic peptide chain release factor subunit 1
|
UniprotKB/SwissProt ID: ERF1_MOUSE (Q8BWY3)
Gene Name: Etf1
Organism: Mus musculus (Mouse).
Function: Directs the termination of nascent peptide synthesis (translation) in response to the termination codons UAA, UAG and UGA (By similarity). Component of the transient SURF complex which recruits UPF1 to stalled ribosomes in the context of nonsense- mediated decay (NMD) of mRNAs containing premature stop codons (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cytoplasm (By similarity).
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 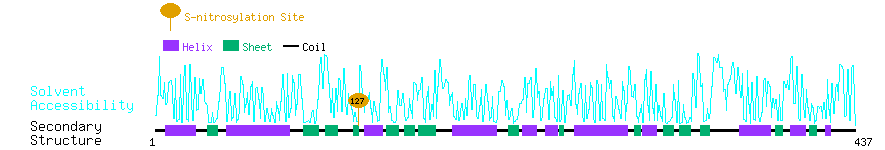 |
|

