Protein Name: Filamin-A
|
UniprotKB/SwissProt ID: FLNA_MOUSE (Q8BTM8)
Gene Name: Flna
Synonyms: Fln, Fln1
Organism: Mus musculus (Mouse).
Function: Promotes orthogonal branching of actin filaments and links actin filaments to membrane glycoproteins. Anchors various transmembrane proteins to the actin cytoskeleton and serves as a scaffold for a wide range of cytoplasmic signaling proteins (By similarity). Interaction with FLNA may allow neuroblast migration from the ventricular zone into the cortical plate. Tethers cell surface-localized furin, modulates its rate of internalization and directs its intracellular trafficking. Involved in ciliogenesis.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cytoplasm, cell cortex (By similarity). Cytoplasm, cytoskeleton (By similarity).
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 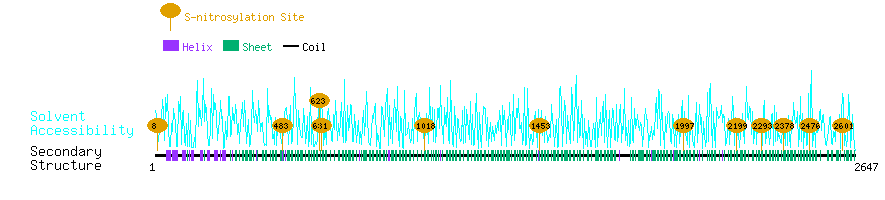 |
|

