Protein Name: Formimidoyltransferase-cyclodeaminase
|
UniprotKB/SwissProt ID: FTCD_MOUSE (Q91XD4)
Gene Name: Ftcd
Organism: Mus musculus (Mouse).
Function: Folate-dependent enzyme, that displays both transferase and deaminase activity. Serves to channel one-carbon units from formiminoglutamate to the folate pool (By similarity). Binds and promotes bundling of vimentin filaments originating from the Golgi (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cytoplasm, cytoskeleton, microtubule organizing center, centrosome, centriole (By similarity). Golgi apparatus (By similarity). Note=More abundantly located around the mother centriole (By similarity).
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 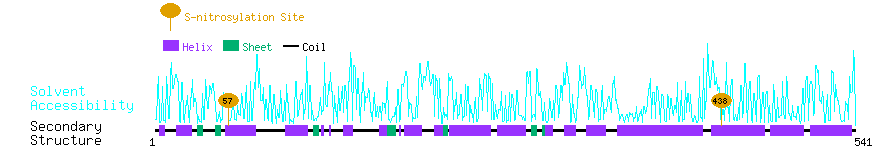 |
|

