Protein Name: Guanine nucleotide-binding protein subunit beta-2-like 1
|
UniprotKB/SwissProt ID: GBLP_HUMAN (P63244)
Gene Name: GNB2L1
Organism: Homo sapiens (Human).
Function: Involved in the recruitment, assembly and/or regulation of a variety of signaling molecules. Interacts with a wide variety of proteins and plays a role in many cellular processes. Component of the 40S ribosomal subunit involved in translational repression. Binds to and stabilizes activated protein kinase C (PKC), increasing PKC-mediated phosphorylation. May recruit activated PKC to the ribosome, leading to phosphorylation of EIF6. Inhibits the activity of SRC kinases including SRC, LCK and YES1. Inhibits cell growth by prolonging the G0/G1 phase of the cell cycle. Enhances phosphorylation of BMAL1 by PRKCA and inhibits transcriptional activity of the BMAL1-CLOCK heterodimer. Facilitates ligand- independent nuclear translocation of AR following PKC activation, represses AR transactivation activity and is required for phosphorylation of AR by SRC. Modulates IGF1R-dependent integrin signaling and promotes cell spreading and contact with the extracellular matrix. Involved in PKC-dependent translocation of ADAM12 to the cell membrane. Promotes the ubiquitination and proteasome-mediated degradation of proteins such as CLEC1B and HIF1A. Required for VANGL2 membrane localization, inhibits Wnt signaling, and regulates cellular polarization and oriented cell division during gastrulation. Required for PTK2/FAK1 phosphorylation and dephosphorylation. Regulates internalization of the muscarinic receptor CHRM2. Promotes apoptosis by increasing oligomerization of BAX and disrupting the interaction of BAX with the anti-apoptotic factor BCL2L. Inhibits TRPM6 channel activity. Regulates cell surface expression of some GPCRs such as TBXA2R. Plays a role in regulation of FLT1-mediated cell migration. Binds to Y.pseudotuberculosis yopK which leads to inhibition of phagocytosis and survival of bacteria following infection of host cells. Enhances phosphorylation of HIV-1 Nef by PKCs. Promotes migration of breast carcinoma cells by binding to and activating RHOA.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cell membrane; Peripheral membrane protein. Cytoplasm. Cytoplasm, perinuclear region. Cytoplasm, cytoskeleton. Nucleus. Perikaryon (By similarity). Cell projection, dendrite (By similarity). Cell projection, phagocytic cup. Note=Recruited to the plasma m
| |
|
Protein disease:
|
| Disease database |
Database Entry |
Disease information | | OMIM | 176981 | GUANINE NUCLEOTIDE-BINDING PROTEIN, BETA-2-LIKE 1; GNB2L1 ;;PROTEIN KINASE C, RECEPTOR FOR A |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 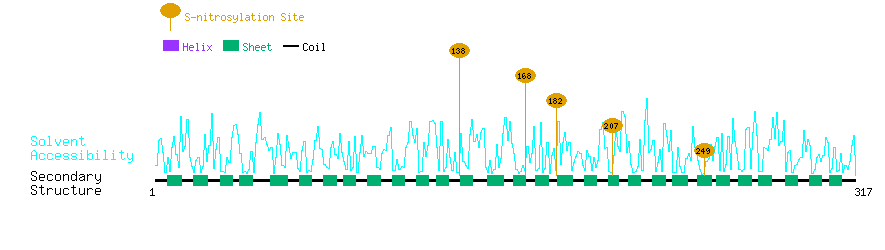 |
|
