Protein Name: Heat shock protein HSP 90-beta
|
UniprotKB/SwissProt ID: HS90B_MOUSE (P11499)
Gene Name: Hsp90ab1
Synonyms: Hsp84, Hsp84-1, Hspcb
Organism: Mus musculus (Mouse).
Function: Molecular chaperone that promotes the maturation, structural maintenance and proper regulation of specific target proteins involved for instance in cell cycle control and signal transduction. Undergoes a functional cycle that is linked to its ATPase activity. This cycle probably induces conformational changes in the client proteins, thereby causing their activation. Interacts dynamically with various co-chaperones that modulate its substrate recognition, ATPase cycle and chaperone function (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cytoplasm. Melanosome (By similarity).
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 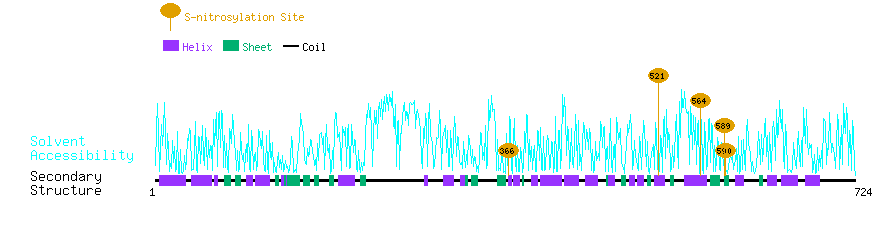 |
|

